Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
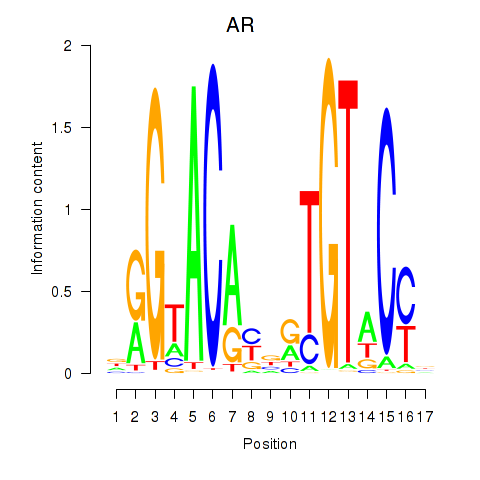
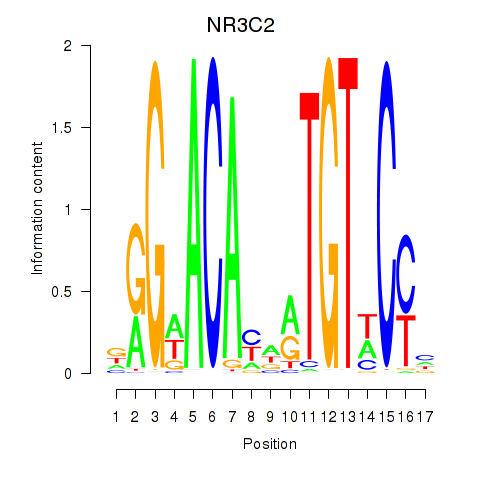
Results for AR_NR3C2
Z-value: 0.57
Transcription factors associated with AR_NR3C2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
AR
|
ENSG00000169083.11 | androgen receptor |
|
NR3C2
|
ENSG00000151623.10 | nuclear receptor subfamily 3 group C member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR3C2 | hg19_v2_chr4_-_149363376_149363492 | 0.32 | 5.3e-01 | Click! |
| AR | hg19_v2_chrX_+_66764375_66764465 | 0.10 | 8.5e-01 | Click! |
Activity profile of AR_NR3C2 motif
Sorted Z-values of AR_NR3C2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_26235206 | 0.70 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr16_-_787771 | 0.40 |
ENST00000568545.1
|
NARFL
|
nuclear prelamin A recognition factor-like |
| chr1_+_1846519 | 0.38 |
ENST00000378604.3
|
CALML6
|
calmodulin-like 6 |
| chr17_+_79071365 | 0.37 |
ENST00000576756.1
|
BAIAP2
|
BAI1-associated protein 2 |
| chr16_-_787728 | 0.32 |
ENST00000567403.1
ENST00000562421.1 |
NARFL
|
nuclear prelamin A recognition factor-like |
| chr5_-_149792295 | 0.31 |
ENST00000518797.1
ENST00000524315.1 ENST00000009530.7 ENST00000377795.3 |
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain |
| chr15_+_31658349 | 0.26 |
ENST00000558844.1
|
KLF13
|
Kruppel-like factor 13 |
| chr17_+_79031415 | 0.26 |
ENST00000572073.1
ENST00000573677.1 |
BAIAP2
|
BAI1-associated protein 2 |
| chr19_+_35630344 | 0.26 |
ENST00000455515.2
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr5_-_141392538 | 0.23 |
ENST00000503794.1
ENST00000510194.1 ENST00000504424.1 ENST00000513454.1 ENST00000458112.2 ENST00000542860.1 ENST00000503229.1 ENST00000500692.2 ENST00000311337.6 ENST00000504139.1 ENST00000505689.1 |
GNPDA1
|
glucosamine-6-phosphate deaminase 1 |
| chr11_-_236326 | 0.23 |
ENST00000525237.1
ENST00000532956.1 ENST00000525319.1 ENST00000524564.1 ENST00000382743.4 |
SIRT3
|
sirtuin 3 |
| chr2_+_238395879 | 0.23 |
ENST00000445024.2
ENST00000338530.4 ENST00000409373.1 |
MLPH
|
melanophilin |
| chr2_+_102953608 | 0.22 |
ENST00000311734.2
ENST00000409584.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr5_+_140186647 | 0.22 |
ENST00000512229.2
ENST00000356878.4 ENST00000530339.1 |
PCDHA4
|
protocadherin alpha 4 |
| chr3_+_125985620 | 0.21 |
ENST00000511512.1
ENST00000512435.1 |
RP11-71E19.1
|
RP11-71E19.1 |
| chr17_-_19290117 | 0.20 |
ENST00000497081.2
|
MFAP4
|
microfibrillar-associated protein 4 |
| chr2_+_238395803 | 0.20 |
ENST00000264605.3
|
MLPH
|
melanophilin |
| chr2_-_27435634 | 0.20 |
ENST00000430186.1
|
SLC5A6
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr2_-_27435390 | 0.19 |
ENST00000428518.1
|
SLC5A6
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr17_-_79304150 | 0.19 |
ENST00000574093.1
|
TMEM105
|
transmembrane protein 105 |
| chr9_-_35563896 | 0.18 |
ENST00000399742.2
|
FAM166B
|
family with sequence similarity 166, member B |
| chr12_+_122241928 | 0.17 |
ENST00000604567.1
ENST00000542440.1 |
SETD1B
|
SET domain containing 1B |
| chr2_-_242576864 | 0.16 |
ENST00000407315.1
|
THAP4
|
THAP domain containing 4 |
| chr17_-_19290483 | 0.16 |
ENST00000395592.2
ENST00000299610.4 |
MFAP4
|
microfibrillar-associated protein 4 |
| chr1_-_1149506 | 0.15 |
ENST00000379236.3
|
TNFRSF4
|
tumor necrosis factor receptor superfamily, member 4 |
| chr8_+_9046503 | 0.14 |
ENST00000512942.2
|
RP11-10A14.5
|
RP11-10A14.5 |
| chr12_-_11002063 | 0.14 |
ENST00000544994.1
ENST00000228811.4 ENST00000540107.1 |
PRR4
|
proline rich 4 (lacrimal) |
| chr19_+_56187987 | 0.14 |
ENST00000411543.2
|
EPN1
|
epsin 1 |
| chr1_-_145715565 | 0.14 |
ENST00000369288.2
ENST00000369290.1 ENST00000401557.3 |
CD160
|
CD160 molecule |
| chr14_+_32414059 | 0.13 |
ENST00000553330.1
|
RP11-187E13.1
|
Uncharacterized protein |
| chr15_+_65204075 | 0.13 |
ENST00000380230.3
ENST00000357698.3 ENST00000395720.1 ENST00000496660.1 ENST00000319580.8 |
ANKDD1A
|
ankyrin repeat and death domain containing 1A |
| chr1_-_10856694 | 0.13 |
ENST00000377022.3
ENST00000344008.5 |
CASZ1
|
castor zinc finger 1 |
| chr8_-_145634731 | 0.13 |
ENST00000349769.3
|
CPSF1
|
cleavage and polyadenylation specific factor 1, 160kDa |
| chr17_-_3794021 | 0.13 |
ENST00000381769.2
|
CAMKK1
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha |
| chr12_-_106480587 | 0.13 |
ENST00000548902.1
|
NUAK1
|
NUAK family, SNF1-like kinase, 1 |
| chr3_+_52279737 | 0.12 |
ENST00000457351.2
|
PPM1M
|
protein phosphatase, Mg2+/Mn2+ dependent, 1M |
| chr9_+_131683174 | 0.11 |
ENST00000372592.3
ENST00000428610.1 |
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr2_-_74875432 | 0.11 |
ENST00000536235.1
ENST00000421985.1 |
M1AP
|
meiosis 1 associated protein |
| chr16_-_4852915 | 0.11 |
ENST00000322048.7
|
ROGDI
|
rogdi homolog (Drosophila) |
| chr8_-_145634666 | 0.11 |
ENST00000531042.1
|
CPSF1
|
cleavage and polyadenylation specific factor 1, 160kDa |
| chr1_-_204119009 | 0.11 |
ENST00000444817.1
|
ETNK2
|
ethanolamine kinase 2 |
| chr8_-_72756667 | 0.10 |
ENST00000325509.4
|
MSC
|
musculin |
| chr5_-_68339648 | 0.10 |
ENST00000479830.2
|
CTC-498J12.1
|
CTC-498J12.1 |
| chr4_-_8442438 | 0.10 |
ENST00000356406.5
ENST00000413009.2 |
ACOX3
|
acyl-CoA oxidase 3, pristanoyl |
| chr12_+_51318513 | 0.10 |
ENST00000332160.4
|
METTL7A
|
methyltransferase like 7A |
| chr8_+_38614754 | 0.10 |
ENST00000521642.1
|
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr16_-_67978016 | 0.10 |
ENST00000264005.5
|
LCAT
|
lecithin-cholesterol acyltransferase |
| chr18_-_48346298 | 0.10 |
ENST00000398439.3
|
MRO
|
maestro |
| chr19_+_57874835 | 0.09 |
ENST00000543226.1
ENST00000596755.1 ENST00000282282.3 ENST00000597658.1 |
TRAPPC2P1
ZNF547
AC003002.4
|
trafficking protein particle complex 2 pseudogene 1 zinc finger protein 547 Uncharacterized protein |
| chr12_-_53575120 | 0.09 |
ENST00000542115.1
|
CSAD
|
cysteine sulfinic acid decarboxylase |
| chr12_-_56848426 | 0.09 |
ENST00000257979.4
|
MIP
|
major intrinsic protein of lens fiber |
| chr11_-_506739 | 0.09 |
ENST00000529306.1
ENST00000438658.2 ENST00000527485.1 ENST00000397615.2 ENST00000397614.1 |
RNH1
|
ribonuclease/angiogenin inhibitor 1 |
| chrX_+_2609356 | 0.09 |
ENST00000381180.3
ENST00000449611.1 |
CD99
|
CD99 molecule |
| chr19_+_54704610 | 0.09 |
ENST00000302907.4
|
RPS9
|
ribosomal protein S9 |
| chr19_-_14117074 | 0.09 |
ENST00000588885.1
ENST00000254325.4 |
RFX1
|
regulatory factor X, 1 (influences HLA class II expression) |
| chr8_-_110615669 | 0.09 |
ENST00000533394.1
|
SYBU
|
syntabulin (syntaxin-interacting) |
| chr8_-_23021533 | 0.08 |
ENST00000312584.3
|
TNFRSF10D
|
tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain |
| chr14_-_21493884 | 0.08 |
ENST00000556974.1
ENST00000554419.1 ENST00000298687.5 ENST00000397858.1 ENST00000360463.3 ENST00000350792.3 ENST00000397847.2 |
NDRG2
|
NDRG family member 2 |
| chr2_-_73298802 | 0.08 |
ENST00000411783.1
ENST00000410065.1 ENST00000442582.1 ENST00000272433.2 |
SFXN5
|
sideroflexin 5 |
| chr19_+_35630926 | 0.08 |
ENST00000588081.1
ENST00000589121.1 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr2_-_166702601 | 0.08 |
ENST00000428888.1
|
AC009495.4
|
AC009495.4 |
| chr7_+_74379083 | 0.08 |
ENST00000361825.7
|
GATSL1
|
GATS protein-like 1 |
| chr12_+_53443963 | 0.08 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr10_+_696000 | 0.08 |
ENST00000381489.5
|
PRR26
|
proline rich 26 |
| chr4_+_87928140 | 0.07 |
ENST00000307808.6
|
AFF1
|
AF4/FMR2 family, member 1 |
| chr2_-_75788424 | 0.07 |
ENST00000410071.1
|
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr2_+_242577027 | 0.07 |
ENST00000402096.1
ENST00000404914.3 |
ATG4B
|
autophagy related 4B, cysteine peptidase |
| chrX_+_2609317 | 0.06 |
ENST00000381187.3
ENST00000381184.1 |
CD99
|
CD99 molecule |
| chr1_-_27693349 | 0.06 |
ENST00000374040.3
ENST00000357582.2 ENST00000493901.1 |
MAP3K6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr12_+_125549925 | 0.06 |
ENST00000316519.6
|
AACS
|
acetoacetyl-CoA synthetase |
| chr4_-_69817481 | 0.06 |
ENST00000251566.4
|
UGT2A3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr10_+_695888 | 0.06 |
ENST00000441152.2
|
PRR26
|
proline rich 26 |
| chr4_+_186347388 | 0.06 |
ENST00000511138.1
ENST00000511581.1 |
C4orf47
|
chromosome 4 open reading frame 47 |
| chr19_-_8408139 | 0.06 |
ENST00000330915.3
ENST00000593649.1 ENST00000595639.1 |
KANK3
|
KN motif and ankyrin repeat domains 3 |
| chr12_+_53443680 | 0.06 |
ENST00000314250.6
ENST00000451358.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr17_+_38333263 | 0.06 |
ENST00000456989.2
ENST00000543876.1 ENST00000544503.1 ENST00000264644.6 ENST00000538884.1 |
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr16_+_67207838 | 0.06 |
ENST00000566871.1
ENST00000268605.7 |
NOL3
|
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr6_-_111927062 | 0.06 |
ENST00000359831.4
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr16_+_67207872 | 0.05 |
ENST00000563258.1
ENST00000568146.1 |
NOL3
|
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr22_+_24030321 | 0.05 |
ENST00000401461.1
|
RGL4
|
ral guanine nucleotide dissociation stimulator-like 4 |
| chr12_+_56615761 | 0.05 |
ENST00000447747.1
ENST00000399713.2 |
NABP2
|
nucleic acid binding protein 2 |
| chr1_-_24469602 | 0.05 |
ENST00000270800.1
|
IL22RA1
|
interleukin 22 receptor, alpha 1 |
| chr8_-_49833978 | 0.05 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr16_-_4852616 | 0.05 |
ENST00000591392.1
ENST00000587711.1 |
ROGDI
|
rogdi homolog (Drosophila) |
| chr8_-_21669826 | 0.05 |
ENST00000517328.1
|
GFRA2
|
GDNF family receptor alpha 2 |
| chr17_+_38498594 | 0.05 |
ENST00000394081.3
|
RARA
|
retinoic acid receptor, alpha |
| chr5_-_173043591 | 0.05 |
ENST00000285908.5
ENST00000480951.1 ENST00000311086.4 |
BOD1
|
biorientation of chromosomes in cell division 1 |
| chr2_-_231825668 | 0.05 |
ENST00000392039.2
|
GPR55
|
G protein-coupled receptor 55 |
| chrX_+_2609207 | 0.04 |
ENST00000381192.3
|
CD99
|
CD99 molecule |
| chr17_+_46918925 | 0.04 |
ENST00000502761.1
|
CALCOCO2
|
calcium binding and coiled-coil domain 2 |
| chr17_-_14140166 | 0.04 |
ENST00000420162.2
ENST00000431716.2 |
CDRT15
|
CMT1A duplicated region transcript 15 |
| chr22_+_38054721 | 0.04 |
ENST00000215904.6
|
PDXP
|
pyridoxal (pyridoxine, vitamin B6) phosphatase |
| chr10_+_48359344 | 0.04 |
ENST00000412534.1
ENST00000444585.1 |
ZNF488
|
zinc finger protein 488 |
| chr1_+_162335197 | 0.04 |
ENST00000431696.1
|
RP11-565P22.6
|
Uncharacterized protein |
| chr11_-_75380165 | 0.04 |
ENST00000304771.3
|
MAP6
|
microtubule-associated protein 6 |
| chr12_+_1099675 | 0.04 |
ENST00000545318.2
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr16_-_66584059 | 0.04 |
ENST00000417693.3
ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr11_+_236540 | 0.04 |
ENST00000532097.1
|
PSMD13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr18_+_56530136 | 0.03 |
ENST00000591083.1
|
ZNF532
|
zinc finger protein 532 |
| chr12_+_66582919 | 0.03 |
ENST00000545837.1
ENST00000457197.2 |
IRAK3
|
interleukin-1 receptor-associated kinase 3 |
| chr22_+_19939026 | 0.03 |
ENST00000406520.3
|
COMT
|
catechol-O-methyltransferase |
| chr1_+_76262552 | 0.03 |
ENST00000263187.3
|
MSH4
|
mutS homolog 4 |
| chr12_+_125549973 | 0.03 |
ENST00000536752.1
ENST00000261686.6 |
AACS
|
acetoacetyl-CoA synthetase |
| chr16_+_56642489 | 0.03 |
ENST00000561491.1
|
MT2A
|
metallothionein 2A |
| chr6_+_123317116 | 0.03 |
ENST00000275162.5
|
CLVS2
|
clavesin 2 |
| chr4_+_37892682 | 0.03 |
ENST00000508802.1
ENST00000261439.4 ENST00000402522.1 |
TBC1D1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| chr9_-_106087316 | 0.03 |
ENST00000411575.1
|
RP11-341A22.2
|
RP11-341A22.2 |
| chr1_-_159832438 | 0.02 |
ENST00000368100.1
|
VSIG8
|
V-set and immunoglobulin domain containing 8 |
| chr11_-_114271139 | 0.02 |
ENST00000325636.4
|
C11orf71
|
chromosome 11 open reading frame 71 |
| chr18_-_48346415 | 0.02 |
ENST00000431965.2
ENST00000436348.2 |
MRO
|
maestro |
| chr4_+_8442496 | 0.02 |
ENST00000389737.4
ENST00000504134.1 |
TRMT44
|
tRNA methyltransferase 44 homolog (S. cerevisiae) |
| chr19_+_1261106 | 0.02 |
ENST00000588411.1
|
CIRBP
|
cold inducible RNA binding protein |
| chr10_-_87551311 | 0.02 |
ENST00000536331.1
|
GRID1
|
glutamate receptor, ionotropic, delta 1 |
| chr2_-_109605663 | 0.02 |
ENST00000409271.1
ENST00000258443.2 ENST00000376651.1 |
EDAR
|
ectodysplasin A receptor |
| chr19_-_45657028 | 0.02 |
ENST00000429338.1
ENST00000589776.1 |
NKPD1
|
NTPase, KAP family P-loop domain containing 1 |
| chr2_+_242577097 | 0.02 |
ENST00000419606.1
ENST00000474739.2 ENST00000396411.3 ENST00000425239.1 ENST00000400771.3 ENST00000430617.2 |
ATG4B
|
autophagy related 4B, cysteine peptidase |
| chr18_-_48346130 | 0.02 |
ENST00000592966.1
|
MRO
|
maestro |
| chr16_+_56672571 | 0.02 |
ENST00000290705.8
|
MT1A
|
metallothionein 1A |
| chr8_-_49834299 | 0.02 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2 |
| chr4_-_135122903 | 0.02 |
ENST00000421491.3
ENST00000529122.2 |
PABPC4L
|
poly(A) binding protein, cytoplasmic 4-like |
| chr19_-_42916499 | 0.02 |
ENST00000601189.1
ENST00000599211.1 |
LIPE
|
lipase, hormone-sensitive |
| chr19_+_58570605 | 0.02 |
ENST00000359978.6
ENST00000401053.4 ENST00000439855.2 ENST00000313434.5 ENST00000511556.1 ENST00000506786.1 |
ZNF135
|
zinc finger protein 135 |
| chr13_+_113777105 | 0.02 |
ENST00000409306.1
ENST00000375551.3 ENST00000375559.3 |
F10
|
coagulation factor X |
| chr19_+_35630628 | 0.02 |
ENST00000588715.1
ENST00000588607.1 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr2_-_37551846 | 0.01 |
ENST00000443187.1
|
PRKD3
|
protein kinase D3 |
| chrX_+_153029633 | 0.01 |
ENST00000538966.1
ENST00000361971.5 ENST00000538776.1 ENST00000538543.1 |
PLXNB3
|
plexin B3 |
| chr19_-_14945933 | 0.01 |
ENST00000322301.3
|
OR7A5
|
olfactory receptor, family 7, subfamily A, member 5 |
| chr3_+_13521665 | 0.01 |
ENST00000295757.3
ENST00000402259.1 ENST00000402271.1 ENST00000446613.2 ENST00000404548.1 ENST00000404040.1 |
HDAC11
|
histone deacetylase 11 |
| chr17_-_65242063 | 0.01 |
ENST00000581159.1
|
HELZ
|
helicase with zinc finger |
| chr2_+_27435734 | 0.01 |
ENST00000419744.1
|
ATRAID
|
all-trans retinoic acid-induced differentiation factor |
| chr1_+_93811438 | 0.01 |
ENST00000370272.4
ENST00000370267.1 |
DR1
|
down-regulator of transcription 1, TBP-binding (negative cofactor 2) |
| chr16_+_56642041 | 0.01 |
ENST00000245185.5
|
MT2A
|
metallothionein 2A |
| chr7_-_4877677 | 0.01 |
ENST00000538469.1
|
RADIL
|
Ras association and DIL domains |
| chrX_-_117119243 | 0.01 |
ENST00000539496.1
ENST00000469946.1 |
KLHL13
|
kelch-like family member 13 |
| chrX_-_80377118 | 0.01 |
ENST00000373250.3
|
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr20_+_15177480 | 0.00 |
ENST00000402914.1
|
MACROD2
|
MACRO domain containing 2 |
| chr3_-_24207039 | 0.00 |
ENST00000280696.5
|
THRB
|
thyroid hormone receptor, beta |
| chr15_-_28344439 | 0.00 |
ENST00000431101.1
ENST00000445578.1 ENST00000353809.5 ENST00000382996.2 ENST00000354638.3 |
OCA2
|
oculocutaneous albinism II |
Network of associatons between targets according to the STRING database.
First level regulatory network of AR_NR3C2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.2 | GO:0006043 | glucosamine catabolic process(GO:0006043) |
| 0.1 | 0.6 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.1 | 0.4 | GO:0015878 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) |
| 0.1 | 0.3 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.1 | 0.7 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 0.4 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.0 | 0.1 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.0 | 0.1 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.2 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.4 | GO:0071493 | elastic fiber assembly(GO:0048251) cellular response to UV-B(GO:0071493) |
| 0.0 | 0.4 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.1 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.1 | GO:0090107 | regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.0 | 0.3 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.0 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.2 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.3 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.7 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.1 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 0.2 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.4 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.1 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.7 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0035877 | death effector domain binding(GO:0035877) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |