Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
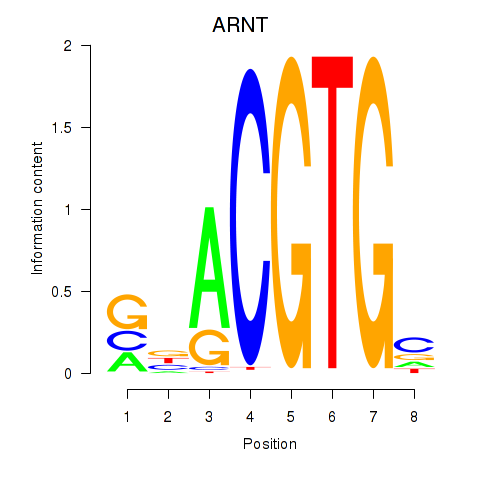
Results for ARNT
Z-value: 0.78
Transcription factors associated with ARNT
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ARNT
|
ENSG00000143437.16 | aryl hydrocarbon receptor nuclear translocator |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARNT | hg19_v2_chr1_-_150849208_150849244 | 0.23 | 6.6e-01 | Click! |
Activity profile of ARNT motif
Sorted Z-values of ARNT motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_150254936 | 0.76 |
ENST00000447007.1
ENST00000369095.1 ENST00000369094.1 |
C1orf51
|
chromosome 1 open reading frame 51 |
| chr1_+_92495528 | 0.61 |
ENST00000370383.4
|
EPHX4
|
epoxide hydrolase 4 |
| chr19_+_50031547 | 0.58 |
ENST00000597801.1
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr1_-_8939265 | 0.47 |
ENST00000489867.1
|
ENO1
|
enolase 1, (alpha) |
| chr19_-_17622269 | 0.47 |
ENST00000595116.1
|
CTD-3131K8.2
|
CTD-3131K8.2 |
| chr17_-_55162360 | 0.45 |
ENST00000576871.1
ENST00000576313.1 |
RP11-166P13.3
|
RP11-166P13.3 |
| chr2_+_232575168 | 0.40 |
ENST00000440384.1
|
PTMA
|
prothymosin, alpha |
| chr3_+_186648507 | 0.39 |
ENST00000458216.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr20_-_62493217 | 0.36 |
ENST00000601296.1
|
C20ORF135
|
C20ORF135 |
| chr17_-_8059638 | 0.35 |
ENST00000584202.1
ENST00000354903.5 ENST00000577253.1 |
PER1
|
period circadian clock 1 |
| chr19_-_19739321 | 0.33 |
ENST00000588461.1
|
LPAR2
|
lysophosphatidic acid receptor 2 |
| chr12_+_112451222 | 0.30 |
ENST00000552052.1
|
ERP29
|
endoplasmic reticulum protein 29 |
| chr3_+_100211412 | 0.27 |
ENST00000323523.4
ENST00000403410.1 ENST00000449609.1 |
TMEM45A
|
transmembrane protein 45A |
| chr4_+_17578815 | 0.26 |
ENST00000226299.4
|
LAP3
|
leucine aminopeptidase 3 |
| chr19_+_19516561 | 0.25 |
ENST00000457895.2
|
GATAD2A
|
GATA zinc finger domain containing 2A |
| chr3_-_19988462 | 0.24 |
ENST00000344838.4
|
EFHB
|
EF-hand domain family, member B |
| chr2_+_105471969 | 0.24 |
ENST00000361360.2
|
POU3F3
|
POU class 3 homeobox 3 |
| chr8_+_26240666 | 0.23 |
ENST00000523949.1
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr1_+_100435986 | 0.22 |
ENST00000532693.1
|
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
| chr3_-_52719888 | 0.22 |
ENST00000458294.1
|
PBRM1
|
polybromo 1 |
| chr9_+_37120536 | 0.21 |
ENST00000336755.5
ENST00000534928.1 ENST00000322831.6 |
ZCCHC7
|
zinc finger, CCHC domain containing 7 |
| chr4_+_76649753 | 0.21 |
ENST00000603759.1
|
USO1
|
USO1 vesicle transport factor |
| chr1_+_45140400 | 0.20 |
ENST00000453711.1
|
C1orf228
|
chromosome 1 open reading frame 228 |
| chr12_+_46777450 | 0.20 |
ENST00000551503.1
|
RP11-96H19.1
|
RP11-96H19.1 |
| chr22_+_41777927 | 0.20 |
ENST00000266304.4
|
TEF
|
thyrotrophic embryonic factor |
| chr3_-_128840604 | 0.19 |
ENST00000476465.1
ENST00000315150.5 ENST00000393304.1 ENST00000393308.1 ENST00000393307.1 ENST00000393305.1 |
RAB43
|
RAB43, member RAS oncogene family |
| chr7_+_72848092 | 0.19 |
ENST00000344575.3
|
FZD9
|
frizzled family receptor 9 |
| chr4_-_4291761 | 0.19 |
ENST00000513174.1
|
LYAR
|
Ly1 antibody reactive |
| chr22_-_39548443 | 0.18 |
ENST00000401405.3
|
CBX7
|
chromobox homolog 7 |
| chr15_+_89182178 | 0.18 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr6_-_83775489 | 0.17 |
ENST00000369747.3
|
UBE3D
|
ubiquitin protein ligase E3D |
| chrX_-_119693745 | 0.17 |
ENST00000371323.2
|
CUL4B
|
cullin 4B |
| chr4_-_76649546 | 0.16 |
ENST00000508510.1
ENST00000509561.1 ENST00000499709.2 ENST00000511868.1 |
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr1_-_9189144 | 0.16 |
ENST00000414642.2
|
GPR157
|
G protein-coupled receptor 157 |
| chr1_-_223537475 | 0.16 |
ENST00000344029.6
ENST00000494793.2 ENST00000366878.4 ENST00000366877.3 |
SUSD4
|
sushi domain containing 4 |
| chr8_+_126442563 | 0.16 |
ENST00000311922.3
|
TRIB1
|
tribbles pseudokinase 1 |
| chr3_-_149688971 | 0.16 |
ENST00000498307.1
ENST00000489155.1 |
PFN2
|
profilin 2 |
| chr1_-_92351666 | 0.16 |
ENST00000465892.2
ENST00000417833.2 |
TGFBR3
|
transforming growth factor, beta receptor III |
| chr6_+_31865552 | 0.16 |
ENST00000469372.1
ENST00000497706.1 |
C2
|
complement component 2 |
| chr7_-_27170352 | 0.16 |
ENST00000428284.2
ENST00000360046.5 |
HOXA4
|
homeobox A4 |
| chr5_-_172755056 | 0.15 |
ENST00000520648.1
|
STC2
|
stanniocalcin 2 |
| chr12_-_15374328 | 0.15 |
ENST00000537647.1
|
RERG
|
RAS-like, estrogen-regulated, growth inhibitor |
| chr14_-_62217779 | 0.15 |
ENST00000554254.1
|
HIF1A-AS2
|
HIF1A antisense RNA 2 |
| chr15_+_68346501 | 0.15 |
ENST00000249636.6
|
PIAS1
|
protein inhibitor of activated STAT, 1 |
| chr16_-_71323617 | 0.14 |
ENST00000563876.1
|
CMTR2
|
cap methyltransferase 2 |
| chr17_+_72428266 | 0.14 |
ENST00000582473.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr19_-_663171 | 0.14 |
ENST00000606896.1
ENST00000589762.2 |
RNF126
|
ring finger protein 126 |
| chr10_-_126849626 | 0.14 |
ENST00000530884.1
|
CTBP2
|
C-terminal binding protein 2 |
| chr20_-_44540686 | 0.14 |
ENST00000477313.1
ENST00000542937.1 ENST00000372431.3 ENST00000354050.4 ENST00000420868.2 |
PLTP
|
phospholipid transfer protein |
| chr7_-_130597935 | 0.14 |
ENST00000447307.1
ENST00000418546.1 |
MIR29B1
|
microRNA 29a |
| chr3_-_42306248 | 0.14 |
ENST00000334681.5
|
CCK
|
cholecystokinin |
| chr3_+_32433154 | 0.13 |
ENST00000334983.5
ENST00000349718.4 |
CMTM7
|
CKLF-like MARVEL transmembrane domain containing 7 |
| chr4_+_76649797 | 0.13 |
ENST00000538159.1
ENST00000514213.2 |
USO1
|
USO1 vesicle transport factor |
| chr12_+_54519842 | 0.13 |
ENST00000508564.1
|
RP11-834C11.4
|
RP11-834C11.4 |
| chr10_-_44070016 | 0.13 |
ENST00000374446.2
ENST00000426961.1 ENST00000535642.1 |
ZNF239
|
zinc finger protein 239 |
| chr9_+_103235365 | 0.13 |
ENST00000374879.4
|
TMEFF1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr2_-_96811170 | 0.13 |
ENST00000288943.4
|
DUSP2
|
dual specificity phosphatase 2 |
| chr17_+_7608511 | 0.13 |
ENST00000226091.2
|
EFNB3
|
ephrin-B3 |
| chr2_+_201170999 | 0.13 |
ENST00000439395.1
ENST00000444012.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr20_+_2633269 | 0.13 |
ENST00000445139.1
|
NOP56
|
NOP56 ribonucleoprotein |
| chr3_+_99536663 | 0.13 |
ENST00000421999.2
ENST00000463526.1 |
CMSS1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr1_+_173837488 | 0.13 |
ENST00000427304.1
ENST00000432989.1 ENST00000367702.1 |
ZBTB37
|
zinc finger and BTB domain containing 37 |
| chr14_+_56046990 | 0.13 |
ENST00000438792.2
ENST00000395314.3 ENST00000395308.1 |
KTN1
|
kinectin 1 (kinesin receptor) |
| chr17_-_30470154 | 0.12 |
ENST00000398832.2
|
AC090616.2
|
Uncharacterized protein |
| chr13_+_25875785 | 0.12 |
ENST00000381747.3
|
NUPL1
|
nucleoporin like 1 |
| chr8_+_104383759 | 0.12 |
ENST00000415886.2
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr3_-_170626376 | 0.12 |
ENST00000487522.1
ENST00000474417.1 |
EIF5A2
|
eukaryotic translation initiation factor 5A2 |
| chr2_+_176972000 | 0.12 |
ENST00000249504.5
|
HOXD11
|
homeobox D11 |
| chr2_-_148778323 | 0.12 |
ENST00000440042.1
ENST00000535373.1 ENST00000540442.1 ENST00000536575.1 |
ORC4
|
origin recognition complex, subunit 4 |
| chr7_-_6048702 | 0.12 |
ENST00000265849.7
|
PMS2
|
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| chr1_-_220101944 | 0.12 |
ENST00000366926.3
ENST00000536992.1 |
SLC30A10
|
solute carrier family 30, member 10 |
| chr2_+_187350973 | 0.12 |
ENST00000544130.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr18_+_23806437 | 0.12 |
ENST00000578121.1
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa |
| chr7_+_1084206 | 0.12 |
ENST00000444847.1
|
GPR146
|
G protein-coupled receptor 146 |
| chr5_-_172198190 | 0.12 |
ENST00000239223.3
|
DUSP1
|
dual specificity phosphatase 1 |
| chr19_-_17799135 | 0.12 |
ENST00000552293.1
ENST00000551649.1 ENST00000550896.1 |
UNC13A
|
unc-13 homolog A (C. elegans) |
| chr15_+_41056218 | 0.12 |
ENST00000260447.4
|
GCHFR
|
GTP cyclohydrolase I feedback regulator |
| chr5_-_38845812 | 0.12 |
ENST00000513480.1
ENST00000512519.1 |
CTD-2127H9.1
|
CTD-2127H9.1 |
| chr13_+_103451548 | 0.12 |
ENST00000419638.1
|
BIVM
|
basic, immunoglobulin-like variable motif containing |
| chr14_+_56046914 | 0.11 |
ENST00000413890.2
ENST00000395309.3 ENST00000554567.1 ENST00000555498.1 |
KTN1
|
kinectin 1 (kinesin receptor) |
| chr1_-_31712401 | 0.11 |
ENST00000373736.2
|
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr7_+_26331678 | 0.11 |
ENST00000446848.2
|
SNX10
|
sorting nexin 10 |
| chr19_+_1205740 | 0.11 |
ENST00000326873.7
|
STK11
|
serine/threonine kinase 11 |
| chr18_+_9913977 | 0.11 |
ENST00000400000.2
ENST00000340541.4 |
VAPA
|
VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa |
| chr13_-_31736132 | 0.11 |
ENST00000429785.2
|
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr19_-_55895966 | 0.11 |
ENST00000444469.3
|
TMEM238
|
transmembrane protein 238 |
| chr2_+_187350883 | 0.11 |
ENST00000337859.6
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr11_+_63998006 | 0.11 |
ENST00000355040.4
|
DNAJC4
|
DnaJ (Hsp40) homolog, subfamily C, member 4 |
| chrX_-_129244336 | 0.11 |
ENST00000434609.1
|
ELF4
|
E74-like factor 4 (ets domain transcription factor) |
| chr12_-_120907374 | 0.11 |
ENST00000550458.1
|
SRSF9
|
serine/arginine-rich splicing factor 9 |
| chr15_+_74218787 | 0.11 |
ENST00000261921.7
|
LOXL1
|
lysyl oxidase-like 1 |
| chr6_+_41888926 | 0.11 |
ENST00000230340.4
|
BYSL
|
bystin-like |
| chr17_+_57642886 | 0.11 |
ENST00000251241.4
ENST00000451169.2 ENST00000425628.3 ENST00000584385.1 ENST00000580030.1 |
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr22_-_42310570 | 0.10 |
ENST00000457093.1
|
SHISA8
|
shisa family member 8 |
| chr15_+_89182156 | 0.10 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr11_+_1411129 | 0.10 |
ENST00000308219.9
ENST00000528841.1 ENST00000531197.1 ENST00000308230.5 |
BRSK2
|
BR serine/threonine kinase 2 |
| chr17_-_2615031 | 0.10 |
ENST00000576885.1
ENST00000574426.2 |
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr15_+_52311398 | 0.10 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr1_+_40420802 | 0.10 |
ENST00000372811.5
ENST00000420632.2 ENST00000434861.1 ENST00000372809.5 |
MFSD2A
|
major facilitator superfamily domain containing 2A |
| chr9_+_103204553 | 0.10 |
ENST00000502978.1
ENST00000334943.6 |
MSANTD3-TMEFF1
TMEFF1
|
MSANTD3-TMEFF1 readthrough transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr4_+_129732419 | 0.10 |
ENST00000510308.1
|
PHF17
|
jade family PHD finger 1 |
| chr6_+_30539153 | 0.10 |
ENST00000326195.8
ENST00000376545.3 ENST00000396515.4 ENST00000441867.1 ENST00000468958.1 |
ABCF1
|
ATP-binding cassette, sub-family F (GCN20), member 1 |
| chr2_+_171627597 | 0.10 |
ENST00000429172.1
ENST00000426475.1 |
AC007405.6
|
AC007405.6 |
| chr1_+_74663994 | 0.10 |
ENST00000472069.1
|
FPGT
|
fucose-1-phosphate guanylyltransferase |
| chr7_-_127032114 | 0.10 |
ENST00000436992.1
|
ZNF800
|
zinc finger protein 800 |
| chr10_-_93392811 | 0.10 |
ENST00000238994.5
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C |
| chr12_+_64798095 | 0.10 |
ENST00000332707.5
|
XPOT
|
exportin, tRNA |
| chr19_-_44258770 | 0.10 |
ENST00000601925.1
ENST00000602222.1 ENST00000599804.1 |
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr3_-_149688896 | 0.10 |
ENST00000239940.7
|
PFN2
|
profilin 2 |
| chr2_-_235405168 | 0.10 |
ENST00000339728.3
|
ARL4C
|
ADP-ribosylation factor-like 4C |
| chr6_+_89791507 | 0.10 |
ENST00000354922.3
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr9_-_73029540 | 0.10 |
ENST00000377126.2
|
KLF9
|
Kruppel-like factor 9 |
| chr1_-_166135952 | 0.10 |
ENST00000354422.3
|
FAM78B
|
family with sequence similarity 78, member B |
| chr7_-_127032363 | 0.10 |
ENST00000393312.1
|
ZNF800
|
zinc finger protein 800 |
| chr1_-_166136187 | 0.10 |
ENST00000338353.3
|
FAM78B
|
family with sequence similarity 78, member B |
| chr19_-_10764509 | 0.10 |
ENST00000591501.1
|
ILF3-AS1
|
ILF3 antisense RNA 1 (head to head) |
| chr10_-_103543145 | 0.09 |
ENST00000370110.5
|
NPM3
|
nucleophosmin/nucleoplasmin 3 |
| chr14_+_68286478 | 0.09 |
ENST00000487861.1
|
RAD51B
|
RAD51 paralog B |
| chr14_+_68286516 | 0.09 |
ENST00000471583.1
ENST00000487270.1 ENST00000488612.1 |
RAD51B
|
RAD51 paralog B |
| chr6_-_139695757 | 0.09 |
ENST00000367651.2
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chrX_+_77359726 | 0.09 |
ENST00000442431.1
|
PGK1
|
phosphoglycerate kinase 1 |
| chr2_-_148778258 | 0.09 |
ENST00000392857.5
ENST00000457954.1 ENST00000392858.1 ENST00000542387.1 |
ORC4
|
origin recognition complex, subunit 4 |
| chr18_+_43914159 | 0.09 |
ENST00000588679.1
ENST00000269439.7 ENST00000543885.1 |
RNF165
|
ring finger protein 165 |
| chr11_-_3013482 | 0.09 |
ENST00000529361.1
ENST00000528968.1 ENST00000534372.1 ENST00000531291.1 ENST00000526842.1 |
NAP1L4
|
nucleosome assembly protein 1-like 4 |
| chr8_+_104383728 | 0.09 |
ENST00000330295.5
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr6_+_7107999 | 0.09 |
ENST00000491191.1
ENST00000379938.2 ENST00000471433.1 |
RREB1
|
ras responsive element binding protein 1 |
| chr2_+_201171372 | 0.09 |
ENST00000409140.3
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr2_-_28113217 | 0.09 |
ENST00000444339.2
|
RBKS
|
ribokinase |
| chr9_+_35673853 | 0.09 |
ENST00000378357.4
|
CA9
|
carbonic anhydrase IX |
| chr5_+_67584523 | 0.09 |
ENST00000521409.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr1_+_28261621 | 0.09 |
ENST00000549094.1
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr11_-_6341724 | 0.09 |
ENST00000530979.1
|
PRKCDBP
|
protein kinase C, delta binding protein |
| chr5_+_154238149 | 0.09 |
ENST00000519430.1
ENST00000520671.1 ENST00000521583.1 ENST00000518028.1 ENST00000519404.1 ENST00000519394.1 ENST00000518775.1 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chr1_-_19229218 | 0.09 |
ENST00000432718.1
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr2_+_198365122 | 0.09 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough |
| chr1_+_150229554 | 0.09 |
ENST00000369111.4
|
CA14
|
carbonic anhydrase XIV |
| chrX_+_23685563 | 0.09 |
ENST00000379341.4
|
PRDX4
|
peroxiredoxin 4 |
| chr7_-_105925367 | 0.09 |
ENST00000354289.4
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr11_-_3013497 | 0.09 |
ENST00000448187.1
ENST00000532325.2 ENST00000399614.2 |
NAP1L4
|
nucleosome assembly protein 1-like 4 |
| chr8_+_109455845 | 0.09 |
ENST00000220853.3
|
EMC2
|
ER membrane protein complex subunit 2 |
| chr11_-_3013316 | 0.09 |
ENST00000430811.1
|
NAP1L4
|
nucleosome assembly protein 1-like 4 |
| chr3_+_38179969 | 0.09 |
ENST00000396334.3
ENST00000417037.2 ENST00000424893.1 ENST00000495303.1 ENST00000443433.2 ENST00000421516.1 |
MYD88
|
myeloid differentiation primary response 88 |
| chr2_+_20646824 | 0.09 |
ENST00000272233.4
|
RHOB
|
ras homolog family member B |
| chr17_+_76311791 | 0.09 |
ENST00000586321.1
|
AC061992.2
|
AC061992.2 |
| chr1_-_109203648 | 0.09 |
ENST00000370031.1
|
HENMT1
|
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chr2_+_48010312 | 0.09 |
ENST00000540021.1
|
MSH6
|
mutS homolog 6 |
| chr1_+_6845384 | 0.08 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr2_+_28113583 | 0.08 |
ENST00000344773.2
ENST00000379624.1 ENST00000342045.2 ENST00000379632.2 ENST00000361704.2 |
BRE
|
brain and reproductive organ-expressed (TNFRSF1A modulator) |
| chr20_-_17949100 | 0.08 |
ENST00000431277.1
|
SNX5
|
sorting nexin 5 |
| chr19_-_663277 | 0.08 |
ENST00000292363.5
|
RNF126
|
ring finger protein 126 |
| chr9_+_133569108 | 0.08 |
ENST00000372358.5
ENST00000546165.1 ENST00000372352.3 ENST00000372351.3 ENST00000372350.3 ENST00000495699.2 |
EXOSC2
|
exosome component 2 |
| chr17_+_56833184 | 0.08 |
ENST00000308249.2
|
PPM1E
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr5_+_38845960 | 0.08 |
ENST00000502536.1
|
OSMR
|
oncostatin M receptor |
| chr14_+_65453432 | 0.08 |
ENST00000246166.2
|
FNTB
|
farnesyltransferase, CAAX box, beta |
| chr5_-_131826457 | 0.08 |
ENST00000437654.1
ENST00000245414.4 |
IRF1
|
interferon regulatory factor 1 |
| chr19_-_44258733 | 0.08 |
ENST00000597586.1
ENST00000596714.1 |
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr4_+_95129061 | 0.08 |
ENST00000354268.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr1_+_45140360 | 0.08 |
ENST00000418644.1
ENST00000458657.2 ENST00000441519.1 ENST00000535358.1 ENST00000445071.1 |
C1orf228
|
chromosome 1 open reading frame 228 |
| chr18_+_23806382 | 0.08 |
ENST00000400466.2
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa |
| chr6_+_123110465 | 0.08 |
ENST00000539041.1
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr1_-_86174065 | 0.08 |
ENST00000370574.3
ENST00000431532.2 |
ZNHIT6
|
zinc finger, HIT-type containing 6 |
| chr5_-_37839782 | 0.08 |
ENST00000326524.2
ENST00000515058.1 |
GDNF
|
glial cell derived neurotrophic factor |
| chr6_-_107436473 | 0.08 |
ENST00000369042.1
|
BEND3
|
BEN domain containing 3 |
| chr15_-_60884706 | 0.08 |
ENST00000449337.2
|
RORA
|
RAR-related orphan receptor A |
| chr6_-_163834852 | 0.08 |
ENST00000604200.1
|
CAHM
|
colon adenocarcinoma hypermethylated (non-protein coding) |
| chr3_-_149688502 | 0.08 |
ENST00000481767.1
ENST00000475518.1 |
PFN2
|
profilin 2 |
| chr16_-_1031520 | 0.08 |
ENST00000568394.1
ENST00000565467.1 |
RP11-161M6.2
|
RP11-161M6.2 |
| chr19_-_1237990 | 0.08 |
ENST00000382477.2
ENST00000215376.6 ENST00000590083.1 |
C19orf26
|
chromosome 19 open reading frame 26 |
| chr6_-_114292449 | 0.08 |
ENST00000519065.1
|
HDAC2
|
histone deacetylase 2 |
| chr3_+_5020801 | 0.08 |
ENST00000256495.3
|
BHLHE40
|
basic helix-loop-helix family, member e40 |
| chr19_-_45909585 | 0.08 |
ENST00000593226.1
ENST00000418234.2 |
PPP1R13L
|
protein phosphatase 1, regulatory subunit 13 like |
| chr12_+_6977323 | 0.08 |
ENST00000462761.1
|
TPI1
|
triosephosphate isomerase 1 |
| chr15_+_44084040 | 0.08 |
ENST00000249786.4
|
SERF2
|
small EDRK-rich factor 2 |
| chr7_-_139876734 | 0.08 |
ENST00000006967.5
|
JHDM1D
|
lysine (K)-specific demethylase 7A |
| chr1_-_6269448 | 0.07 |
ENST00000465335.1
|
RPL22
|
ribosomal protein L22 |
| chr2_-_27294500 | 0.07 |
ENST00000447619.1
ENST00000429985.1 ENST00000456793.1 |
OST4
|
oligosaccharyltransferase 4 homolog (S. cerevisiae) |
| chr4_-_57301748 | 0.07 |
ENST00000264220.2
|
PPAT
|
phosphoribosyl pyrophosphate amidotransferase |
| chr4_-_5891918 | 0.07 |
ENST00000512574.1
|
CRMP1
|
collapsin response mediator protein 1 |
| chrX_+_16804544 | 0.07 |
ENST00000380122.5
ENST00000398155.4 |
TXLNG
|
taxilin gamma |
| chr11_-_27722021 | 0.07 |
ENST00000356660.4
ENST00000418212.1 ENST00000533246.1 |
BDNF
|
brain-derived neurotrophic factor |
| chr4_+_95128748 | 0.07 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr6_-_160210604 | 0.07 |
ENST00000420894.2
ENST00000539756.1 ENST00000544255.1 |
TCP1
|
t-complex 1 |
| chr4_+_56212505 | 0.07 |
ENST00000505210.1
|
SRD5A3
|
steroid 5 alpha-reductase 3 |
| chr1_-_45805752 | 0.07 |
ENST00000354383.6
ENST00000355498.2 ENST00000372100.5 ENST00000531105.1 |
MUTYH
|
mutY homolog |
| chr22_+_38864041 | 0.07 |
ENST00000216014.4
ENST00000409006.3 |
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr2_-_171627269 | 0.07 |
ENST00000442456.1
|
AC007405.4
|
AC007405.4 |
| chr3_+_113666748 | 0.07 |
ENST00000330212.3
ENST00000498275.1 |
ZDHHC23
|
zinc finger, DHHC-type containing 23 |
| chr14_+_62162258 | 0.07 |
ENST00000337138.4
ENST00000394997.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chrX_+_129305623 | 0.07 |
ENST00000257017.4
|
RAB33A
|
RAB33A, member RAS oncogene family |
| chr8_-_30515693 | 0.07 |
ENST00000355904.4
|
GTF2E2
|
general transcription factor IIE, polypeptide 2, beta 34kDa |
| chr11_-_118927561 | 0.07 |
ENST00000530473.1
|
HYOU1
|
hypoxia up-regulated 1 |
| chr13_-_36788718 | 0.07 |
ENST00000317764.6
ENST00000379881.3 |
SOHLH2
|
spermatogenesis and oogenesis specific basic helix-loop-helix 2 |
| chr19_-_44285401 | 0.07 |
ENST00000262888.3
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr3_-_145878954 | 0.07 |
ENST00000282903.5
ENST00000360060.3 |
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr2_+_46926048 | 0.07 |
ENST00000306503.5
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr6_-_114292284 | 0.07 |
ENST00000520895.1
ENST00000521163.1 ENST00000524334.1 ENST00000368632.2 ENST00000398283.2 |
HDAC2
|
histone deacetylase 2 |
| chr13_-_31736478 | 0.07 |
ENST00000445273.2
|
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr11_+_66115304 | 0.07 |
ENST00000531602.1
|
RP11-867G23.8
|
Uncharacterized protein |
| chr13_+_52586517 | 0.07 |
ENST00000523764.1
ENST00000521508.1 |
ALG11
|
ALG11, alpha-1,2-mannosyltransferase |
| chr11_+_809647 | 0.07 |
ENST00000321153.4
|
RPLP2
|
ribosomal protein, large, P2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ARNT
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 0.3 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 0.3 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.1 | 0.2 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.2 | GO:0036483 | neuron intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0036483) regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903381) negative regulation of endoplasmic reticulum stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903382) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.3 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.2 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.3 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.2 | GO:0072240 | DCT cell differentiation(GO:0072069) metanephric DCT cell differentiation(GO:0072240) |
| 0.0 | 0.1 | GO:0043095 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.0 | 0.2 | GO:0060979 | transforming growth factor beta receptor complex assembly(GO:0007181) response to luteinizing hormone(GO:0034699) vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.0 | 0.2 | GO:0045658 | eosinophil differentiation(GO:0030222) regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 | 0.2 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.0 | 0.1 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.0 | 0.2 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.1 | GO:0071629 | cytoplasm-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071629) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.2 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.0 | 0.1 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.4 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.1 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.0 | 0.1 | GO:0045590 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.1 | GO:0018277 | protein deamination(GO:0018277) |
| 0.0 | 0.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:1904530 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.1 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.0 | 0.1 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.0 | 0.2 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.1 | GO:0070101 | positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.0 | 0.1 | GO:0035572 | N-terminal protein amino acid methylation(GO:0006480) N-terminal peptidyl-alanine methylation(GO:0018011) N-terminal peptidyl-alanine trimethylation(GO:0018012) N-terminal peptidyl-glycine methylation(GO:0018013) N-terminal peptidyl-proline dimethylation(GO:0018016) peptidyl-alanine modification(GO:0018194) N-terminal peptidyl-proline methylation(GO:0035568) N-terminal peptidyl-serine methylation(GO:0035570) N-terminal peptidyl-serine dimethylation(GO:0035572) N-terminal peptidyl-serine trimethylation(GO:0035573) |
| 0.0 | 0.1 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.1 | GO:1905073 | occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.0 | 0.1 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.1 | GO:0015864 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.1 | GO:0072108 | positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0072108) |
| 0.0 | 0.1 | GO:0035802 | adrenal cortex development(GO:0035801) adrenal cortex formation(GO:0035802) |
| 0.0 | 0.1 | GO:0051808 | translocation of peptides or proteins into host(GO:0042000) translocation of peptides or proteins into host cell cytoplasm(GO:0044053) translocation of molecules into host(GO:0044417) translocation of peptides or proteins into other organism involved in symbiotic interaction(GO:0051808) translocation of molecules into other organism involved in symbiotic interaction(GO:0051836) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.2 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.0 | GO:0009452 | 7-methylguanosine mRNA capping(GO:0006370) 7-methylguanosine RNA capping(GO:0009452) RNA capping(GO:0036260) |
| 0.0 | 0.1 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.0 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.2 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.0 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.0 | 0.0 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.0 | 0.0 | GO:0061461 | lysine import(GO:0034226) L-arginine import(GO:0043091) L-lysine import(GO:0061461) arginine import(GO:0090467) L-lysine import into cell(GO:1903410) |
| 0.0 | 0.1 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.1 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.0 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.0 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.2 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.1 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.1 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.1 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.0 | 0.1 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.1 | GO:0032096 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.0 | 0.0 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.0 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.0 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.0 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.1 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.0 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.1 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.0 | GO:1902356 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.0 | 0.2 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.0 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.0 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0070685 | macropinocytic cup(GO:0070685) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.4 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.0 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.1 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.2 | GO:0070123 | transforming growth factor beta receptor activity, type III(GO:0070123) |
| 0.1 | 0.2 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.1 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
| 0.0 | 0.1 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.1 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 0.0 | 0.1 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.0 | 0.1 | GO:0032408 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.0 | 0.2 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.0 | 0.1 | GO:0070361 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.0 | 0.1 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.2 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.1 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.1 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.2 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.1 | GO:0036009 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.1 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.0 | 0.0 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.0 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.0 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.0 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.0 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.0 | 0.2 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.0 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.7 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |