Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
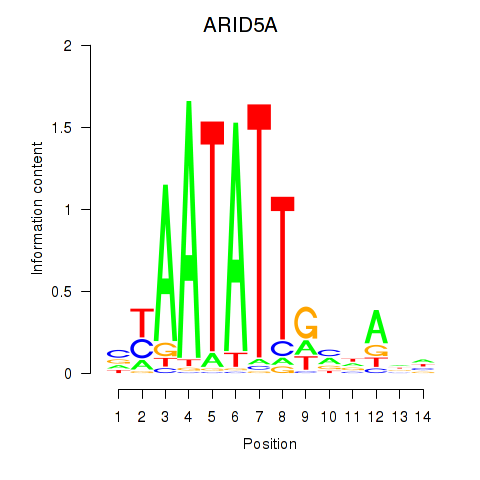
Results for ARID5A
Z-value: 0.96
Transcription factors associated with ARID5A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ARID5A
|
ENSG00000196843.11 | AT-rich interaction domain 5A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARID5A | hg19_v2_chr2_+_97202480_97202499 | 0.09 | 8.7e-01 | Click! |
Activity profile of ARID5A motif
Sorted Z-values of ARID5A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_26104104 | 1.43 |
ENST00000377803.2
|
HIST1H4C
|
histone cluster 1, H4c |
| chr12_+_21207503 | 0.98 |
ENST00000545916.1
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr6_+_26156551 | 0.90 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr12_+_28605426 | 0.66 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr6_-_26235206 | 0.61 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr3_-_150421752 | 0.54 |
ENST00000498386.1
|
FAM194A
|
family with sequence similarity 194, member A |
| chr19_-_58204128 | 0.47 |
ENST00000597520.1
|
AC004017.1
|
Uncharacterized protein |
| chr11_-_74408739 | 0.43 |
ENST00000529912.1
|
CHRDL2
|
chordin-like 2 |
| chr19_-_48753028 | 0.41 |
ENST00000522431.1
|
CARD8
|
caspase recruitment domain family, member 8 |
| chr15_-_54267147 | 0.40 |
ENST00000558866.1
ENST00000558920.1 |
RP11-643A5.2
|
RP11-643A5.2 |
| chr1_+_104159999 | 0.37 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr5_-_157161727 | 0.35 |
ENST00000599823.1
|
AC026407.1
|
Uncharacterized protein |
| chr1_+_948803 | 0.34 |
ENST00000379389.4
|
ISG15
|
ISG15 ubiquitin-like modifier |
| chr6_-_26199499 | 0.33 |
ENST00000377831.5
|
HIST1H3D
|
histone cluster 1, H3d |
| chr15_-_41522889 | 0.32 |
ENST00000458580.2
ENST00000314992.5 ENST00000558396.1 |
EXD1
|
exonuclease 3'-5' domain containing 1 |
| chr19_+_54135310 | 0.32 |
ENST00000376650.1
|
DPRX
|
divergent-paired related homeobox |
| chr11_-_118305921 | 0.30 |
ENST00000532619.1
|
RP11-770J1.4
|
RP11-770J1.4 |
| chr4_-_122148620 | 0.29 |
ENST00000509841.1
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr1_+_43424698 | 0.28 |
ENST00000431759.1
|
SLC2A1-AS1
|
SLC2A1 antisense RNA 1 |
| chr11_-_125366018 | 0.28 |
ENST00000527534.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr18_+_68002675 | 0.27 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr2_+_149447783 | 0.26 |
ENST00000449013.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr2_-_69180083 | 0.26 |
ENST00000328895.4
|
GKN2
|
gastrokine 2 |
| chr17_+_73455788 | 0.26 |
ENST00000581519.1
|
KIAA0195
|
KIAA0195 |
| chr5_-_59668541 | 0.25 |
ENST00000514552.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr21_-_19858196 | 0.24 |
ENST00000422787.1
|
TMPRSS15
|
transmembrane protease, serine 15 |
| chr1_-_179457805 | 0.24 |
ENST00000600581.1
|
AL160286.1
|
Uncharacterized protein |
| chr5_+_140602904 | 0.24 |
ENST00000515856.2
ENST00000239449.4 |
PCDHB14
|
protocadherin beta 14 |
| chr6_+_90272488 | 0.24 |
ENST00000485637.1
ENST00000522705.1 |
ANKRD6
|
ankyrin repeat domain 6 |
| chr4_-_71532339 | 0.23 |
ENST00000254801.4
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr17_-_47925379 | 0.23 |
ENST00000352793.2
ENST00000334568.4 ENST00000398154.1 ENST00000436235.1 ENST00000326219.5 |
TAC4
|
tachykinin 4 (hemokinin) |
| chr14_+_56584414 | 0.23 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr15_-_55657428 | 0.22 |
ENST00000568543.1
|
CCPG1
|
cell cycle progression 1 |
| chr3_-_87325612 | 0.22 |
ENST00000561167.1
ENST00000560656.1 ENST00000344265.3 |
POU1F1
|
POU class 1 homeobox 1 |
| chr8_-_150563 | 0.22 |
ENST00000523795.2
|
RP11-585F1.10
|
Protein LOC100286914 |
| chr6_-_26199471 | 0.22 |
ENST00000341023.1
|
HIST1H2AD
|
histone cluster 1, H2ad |
| chr22_-_23922410 | 0.22 |
ENST00000249053.3
|
IGLL1
|
immunoglobulin lambda-like polypeptide 1 |
| chr17_+_34171081 | 0.22 |
ENST00000585577.1
|
TAF15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa |
| chr10_+_97709725 | 0.22 |
ENST00000472454.2
|
RP11-248J23.6
|
Protein LOC100652732 |
| chr4_+_110834033 | 0.21 |
ENST00000509793.1
ENST00000265171.5 |
EGF
|
epidermal growth factor |
| chrX_-_154255143 | 0.21 |
ENST00000453950.1
ENST00000423959.1 |
F8
|
coagulation factor VIII, procoagulant component |
| chr4_+_71019903 | 0.20 |
ENST00000344526.5
|
C4orf40
|
chromosome 4 open reading frame 40 |
| chr3_-_170744498 | 0.20 |
ENST00000382808.4
ENST00000314251.3 |
SLC2A2
|
solute carrier family 2 (facilitated glucose transporter), member 2 |
| chr20_-_52612468 | 0.20 |
ENST00000422805.1
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr14_+_77244349 | 0.19 |
ENST00000554743.1
|
VASH1
|
vasohibin 1 |
| chr8_-_112039643 | 0.19 |
ENST00000524283.1
|
RP11-946L20.2
|
RP11-946L20.2 |
| chr1_+_79115503 | 0.19 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr20_+_42543504 | 0.19 |
ENST00000341197.4
|
TOX2
|
TOX high mobility group box family member 2 |
| chr6_-_25874440 | 0.19 |
ENST00000361703.6
ENST00000397060.4 |
SLC17A3
|
solute carrier family 17 (organic anion transporter), member 3 |
| chr2_+_183982255 | 0.18 |
ENST00000455063.1
|
NUP35
|
nucleoporin 35kDa |
| chr16_+_55522536 | 0.18 |
ENST00000570283.1
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) |
| chrX_-_107682702 | 0.18 |
ENST00000372216.4
|
COL4A6
|
collagen, type IV, alpha 6 |
| chr5_+_140220769 | 0.18 |
ENST00000531613.1
ENST00000378123.3 |
PCDHA8
|
protocadherin alpha 8 |
| chr5_+_7373230 | 0.18 |
ENST00000500616.2
|
CTD-2296D1.5
|
CTD-2296D1.5 |
| chr10_-_63995871 | 0.17 |
ENST00000315289.2
|
RTKN2
|
rhotekin 2 |
| chrX_-_154493791 | 0.17 |
ENST00000369454.3
|
RAB39B
|
RAB39B, member RAS oncogene family |
| chr4_+_69962185 | 0.17 |
ENST00000305231.7
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr3_-_150421728 | 0.16 |
ENST00000295910.6
ENST00000491361.1 |
FAM194A
|
family with sequence similarity 194, member A |
| chr14_+_39703112 | 0.16 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr17_-_37844267 | 0.16 |
ENST00000579146.1
ENST00000378011.4 ENST00000429199.2 ENST00000300658.4 |
PGAP3
|
post-GPI attachment to proteins 3 |
| chr6_+_123317116 | 0.16 |
ENST00000275162.5
|
CLVS2
|
clavesin 2 |
| chr12_+_32260085 | 0.16 |
ENST00000548411.1
ENST00000281474.5 ENST00000551086.1 |
BICD1
|
bicaudal D homolog 1 (Drosophila) |
| chr4_-_71532207 | 0.16 |
ENST00000543780.1
ENST00000391614.3 |
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr1_-_89531041 | 0.16 |
ENST00000370473.4
|
GBP1
|
guanylate binding protein 1, interferon-inducible |
| chr6_-_169654139 | 0.16 |
ENST00000366787.3
|
THBS2
|
thrombospondin 2 |
| chr22_+_25615489 | 0.16 |
ENST00000398215.2
|
CRYBB2
|
crystallin, beta B2 |
| chr10_+_78078088 | 0.15 |
ENST00000496424.2
|
C10orf11
|
chromosome 10 open reading frame 11 |
| chr17_+_61473104 | 0.15 |
ENST00000583016.1
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr2_+_33661382 | 0.15 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr2_-_118943930 | 0.15 |
ENST00000449075.1
ENST00000414886.1 ENST00000449819.1 |
AC093901.1
|
AC093901.1 |
| chr6_+_63921399 | 0.15 |
ENST00000356170.3
|
FKBP1C
|
FK506 binding protein 1C |
| chr5_+_159848854 | 0.15 |
ENST00000517480.1
ENST00000520452.1 ENST00000393964.1 |
PTTG1
|
pituitary tumor-transforming 1 |
| chr7_+_116593536 | 0.15 |
ENST00000417919.1
|
ST7
|
suppression of tumorigenicity 7 |
| chr6_+_76599809 | 0.15 |
ENST00000430435.1
|
MYO6
|
myosin VI |
| chr1_+_160336851 | 0.14 |
ENST00000302101.5
|
NHLH1
|
nescient helix loop helix 1 |
| chr6_+_90192974 | 0.14 |
ENST00000520458.1
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr22_-_23922448 | 0.14 |
ENST00000438703.1
ENST00000330377.2 |
IGLL1
|
immunoglobulin lambda-like polypeptide 1 |
| chr8_+_67104323 | 0.13 |
ENST00000518412.1
ENST00000518035.1 ENST00000517725.1 |
LINC00967
|
long intergenic non-protein coding RNA 967 |
| chr11_-_6633799 | 0.13 |
ENST00000299424.4
|
TAF10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa |
| chr12_-_70093111 | 0.13 |
ENST00000548658.1
ENST00000476098.1 ENST00000331471.4 ENST00000393365.1 |
BEST3
|
bestrophin 3 |
| chr4_+_100737954 | 0.13 |
ENST00000296414.7
ENST00000512369.1 |
DAPP1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides |
| chr12_-_57006882 | 0.13 |
ENST00000551996.1
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr19_-_893200 | 0.13 |
ENST00000269814.4
ENST00000395808.3 ENST00000312090.6 ENST00000325464.1 |
MED16
|
mediator complex subunit 16 |
| chr7_+_95115210 | 0.13 |
ENST00000428113.1
ENST00000325885.5 |
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr5_-_135290651 | 0.13 |
ENST00000522943.1
ENST00000514447.2 |
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr1_-_154600421 | 0.13 |
ENST00000368471.3
ENST00000292205.5 |
ADAR
|
adenosine deaminase, RNA-specific |
| chr10_+_114710516 | 0.13 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr12_+_12938541 | 0.13 |
ENST00000356591.4
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr12_-_7596735 | 0.12 |
ENST00000416109.2
ENST00000396630.1 ENST00000313599.3 |
CD163L1
|
CD163 molecule-like 1 |
| chr12_+_58087738 | 0.12 |
ENST00000552285.1
|
OS9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin |
| chr12_-_53297432 | 0.12 |
ENST00000546900.1
|
KRT8
|
keratin 8 |
| chr1_-_197036364 | 0.12 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chrX_+_48755183 | 0.12 |
ENST00000376563.1
ENST00000376566.4 |
PQBP1
|
polyglutamine binding protein 1 |
| chr15_-_100258029 | 0.12 |
ENST00000378904.2
|
DKFZP779J2370
|
DKFZP779J2370 |
| chr14_-_92247032 | 0.12 |
ENST00000556661.1
ENST00000553676.1 ENST00000554560.1 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr16_-_48281444 | 0.12 |
ENST00000537808.1
ENST00000569991.1 |
ABCC11
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 11 |
| chr5_-_135290705 | 0.12 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr11_-_73587719 | 0.12 |
ENST00000545127.1
ENST00000537289.1 ENST00000355693.4 |
COA4
|
cytochrome c oxidase assembly factor 4 homolog (S. cerevisiae) |
| chr10_+_114710425 | 0.12 |
ENST00000352065.5
ENST00000369395.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chrX_-_55208866 | 0.12 |
ENST00000545075.1
|
MTRNR2L10
|
MT-RNR2-like 10 |
| chr16_-_4588762 | 0.12 |
ENST00000562334.1
ENST00000562579.1 ENST00000567695.1 ENST00000563507.1 |
CDIP1
|
cell death-inducing p53 target 1 |
| chr1_-_157014865 | 0.12 |
ENST00000361409.2
|
ARHGEF11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr5_-_55412774 | 0.12 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr16_+_69333585 | 0.12 |
ENST00000570054.2
|
RP11-343C2.11
|
Uncharacterized protein |
| chr5_+_159848807 | 0.11 |
ENST00000352433.5
|
PTTG1
|
pituitary tumor-transforming 1 |
| chr5_+_61874696 | 0.11 |
ENST00000491184.2
|
LRRC70
|
leucine rich repeat containing 70 |
| chr12_+_58087901 | 0.11 |
ENST00000315970.7
ENST00000547079.1 ENST00000439210.2 ENST00000389146.6 ENST00000413095.2 ENST00000551035.1 ENST00000257966.8 ENST00000435406.2 ENST00000550372.1 ENST00000389142.5 |
OS9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin |
| chr2_+_223725723 | 0.11 |
ENST00000535678.1
|
ACSL3
|
acyl-CoA synthetase long-chain family member 3 |
| chr10_+_135204338 | 0.11 |
ENST00000468317.2
|
RP11-108K14.8
|
Mitochondrial GTPase 1 |
| chr4_+_76871883 | 0.11 |
ENST00000599764.1
|
AC110615.1
|
Uncharacterized protein |
| chr16_-_4588822 | 0.11 |
ENST00000564828.1
|
CDIP1
|
cell death-inducing p53 target 1 |
| chr18_-_53253112 | 0.11 |
ENST00000568673.1
ENST00000562847.1 ENST00000568147.1 |
TCF4
|
transcription factor 4 |
| chr2_-_188312971 | 0.11 |
ENST00000410068.1
ENST00000447403.1 ENST00000410102.1 |
CALCRL
|
calcitonin receptor-like |
| chr10_-_52645416 | 0.10 |
ENST00000374001.2
ENST00000373997.3 ENST00000373995.3 ENST00000282641.2 ENST00000395495.1 ENST00000414883.1 |
A1CF
|
APOBEC1 complementation factor |
| chr1_+_77333117 | 0.10 |
ENST00000477717.1
|
ST6GALNAC5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr6_-_18249971 | 0.10 |
ENST00000507591.1
|
DEK
|
DEK oncogene |
| chr18_-_67623906 | 0.10 |
ENST00000583955.1
|
CD226
|
CD226 molecule |
| chr3_+_101546827 | 0.10 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr11_-_62477041 | 0.10 |
ENST00000433053.1
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr1_-_89591749 | 0.10 |
ENST00000370466.3
|
GBP2
|
guanylate binding protein 2, interferon-inducible |
| chr17_+_37844331 | 0.10 |
ENST00000578199.1
ENST00000406381.2 |
ERBB2
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr7_-_120498357 | 0.10 |
ENST00000415871.1
ENST00000222747.3 ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr14_-_67878917 | 0.10 |
ENST00000216446.4
|
PLEK2
|
pleckstrin 2 |
| chr9_-_117853297 | 0.10 |
ENST00000542877.1
ENST00000537320.1 ENST00000341037.4 |
TNC
|
tenascin C |
| chr17_+_48624450 | 0.10 |
ENST00000006658.6
ENST00000356488.4 ENST00000393244.3 |
SPATA20
|
spermatogenesis associated 20 |
| chr20_+_30795664 | 0.10 |
ENST00000375749.3
ENST00000375730.3 ENST00000539210.1 |
POFUT1
|
protein O-fucosyltransferase 1 |
| chr2_-_99917639 | 0.10 |
ENST00000308528.4
|
LYG1
|
lysozyme G-like 1 |
| chr1_+_41448820 | 0.10 |
ENST00000372616.1
|
CTPS1
|
CTP synthase 1 |
| chr10_-_127505167 | 0.10 |
ENST00000368786.1
|
UROS
|
uroporphyrinogen III synthase |
| chr3_+_184018352 | 0.10 |
ENST00000435761.1
ENST00000439383.1 |
PSMD2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| chr1_+_160175166 | 0.10 |
ENST00000368077.1
|
PEA15
|
phosphoprotein enriched in astrocytes 15 |
| chr4_+_106631966 | 0.10 |
ENST00000360505.5
ENST00000510865.1 ENST00000509336.1 |
GSTCD
|
glutathione S-transferase, C-terminal domain containing |
| chr7_+_44663908 | 0.10 |
ENST00000543843.1
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr12_-_11287243 | 0.09 |
ENST00000539585.1
|
TAS2R30
|
taste receptor, type 2, member 30 |
| chr1_-_146054494 | 0.09 |
ENST00000401009.2
|
NBPF11
|
neuroblastoma breakpoint family, member 11 |
| chr1_-_113253977 | 0.09 |
ENST00000606505.1
ENST00000605933.1 |
RP11-426L16.10
|
Rho-related GTP-binding protein RhoC |
| chr19_+_35417798 | 0.09 |
ENST00000303586.7
ENST00000439785.1 ENST00000601540.1 |
ZNF30
|
zinc finger protein 30 |
| chr5_-_43397184 | 0.09 |
ENST00000513525.1
|
CCL28
|
chemokine (C-C motif) ligand 28 |
| chr4_-_120222076 | 0.09 |
ENST00000504110.1
|
C4orf3
|
chromosome 4 open reading frame 3 |
| chr21_-_22175450 | 0.09 |
ENST00000435279.2
|
LINC00320
|
long intergenic non-protein coding RNA 320 |
| chr1_-_158656488 | 0.09 |
ENST00000368147.4
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chr3_-_106959424 | 0.09 |
ENST00000607801.1
ENST00000479612.2 ENST00000484698.1 ENST00000477210.2 ENST00000473636.1 |
LINC00882
|
long intergenic non-protein coding RNA 882 |
| chr18_+_39766626 | 0.09 |
ENST00000593234.1
ENST00000585627.1 ENST00000591199.1 ENST00000586990.1 ENST00000593051.1 ENST00000593316.1 ENST00000591381.1 ENST00000585639.1 ENST00000589068.1 |
LINC00907
|
long intergenic non-protein coding RNA 907 |
| chr7_-_86595190 | 0.09 |
ENST00000398276.2
ENST00000416314.1 ENST00000425689.1 |
KIAA1324L
|
KIAA1324-like |
| chr4_-_69536346 | 0.09 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chrX_+_27608490 | 0.09 |
ENST00000451261.2
|
DCAF8L2
|
DDB1 and CUL4 associated factor 8-like 2 |
| chr17_+_53342311 | 0.09 |
ENST00000226067.5
|
HLF
|
hepatic leukemia factor |
| chr4_+_69962212 | 0.09 |
ENST00000508661.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr7_-_45957011 | 0.09 |
ENST00000417621.1
|
IGFBP3
|
insulin-like growth factor binding protein 3 |
| chr6_+_26087646 | 0.09 |
ENST00000309234.6
|
HFE
|
hemochromatosis |
| chr5_+_94982435 | 0.09 |
ENST00000511684.1
ENST00000380005.4 |
RFESD
|
Rieske (Fe-S) domain containing |
| chr6_-_49712147 | 0.09 |
ENST00000433368.2
ENST00000354620.4 |
CRISP3
|
cysteine-rich secretory protein 3 |
| chr4_+_187148556 | 0.09 |
ENST00000264690.6
ENST00000446598.2 ENST00000414291.1 ENST00000513864.1 |
KLKB1
|
kallikrein B, plasma (Fletcher factor) 1 |
| chrX_+_10031499 | 0.09 |
ENST00000454666.1
|
WWC3
|
WWC family member 3 |
| chr7_+_148892557 | 0.08 |
ENST00000262085.3
|
ZNF282
|
zinc finger protein 282 |
| chr16_-_3422283 | 0.08 |
ENST00000399974.3
|
MTRNR2L4
|
MT-RNR2-like 4 |
| chrX_-_134186144 | 0.08 |
ENST00000370775.2
|
FAM127B
|
family with sequence similarity 127, member B |
| chr7_+_16700806 | 0.08 |
ENST00000446596.1
ENST00000438834.1 |
BZW2
|
basic leucine zipper and W2 domains 2 |
| chr16_+_446713 | 0.08 |
ENST00000397722.1
ENST00000454619.1 |
NME4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr12_+_47610315 | 0.08 |
ENST00000548348.1
ENST00000549500.1 |
PCED1B
|
PC-esterase domain containing 1B |
| chr12_+_8995832 | 0.08 |
ENST00000541459.1
|
A2ML1
|
alpha-2-macroglobulin-like 1 |
| chrX_+_48755202 | 0.08 |
ENST00000447146.2
ENST00000376548.5 ENST00000247140.4 |
PQBP1
|
polyglutamine binding protein 1 |
| chr17_+_78518617 | 0.08 |
ENST00000537330.1
ENST00000570891.1 |
RPTOR
|
regulatory associated protein of MTOR, complex 1 |
| chr6_+_32812568 | 0.08 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr1_+_74701062 | 0.08 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr4_+_55095264 | 0.08 |
ENST00000257290.5
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr12_-_10588539 | 0.08 |
ENST00000381902.2
ENST00000381901.1 ENST00000539033.1 |
KLRC2
NKG2-E
|
killer cell lectin-like receptor subfamily C, member 2 Uncharacterized protein |
| chr5_+_140165876 | 0.08 |
ENST00000504120.2
ENST00000394633.3 ENST00000378133.3 |
PCDHA1
|
protocadherin alpha 1 |
| chr2_+_133874577 | 0.08 |
ENST00000596384.1
|
AC011755.1
|
HCG2006742; Protein LOC100996685 |
| chr8_-_52721975 | 0.08 |
ENST00000356297.4
ENST00000543296.1 |
PXDNL
|
peroxidasin homolog (Drosophila)-like |
| chr8_-_59412717 | 0.08 |
ENST00000301645.3
|
CYP7A1
|
cytochrome P450, family 7, subfamily A, polypeptide 1 |
| chr7_-_45956856 | 0.08 |
ENST00000428530.1
|
IGFBP3
|
insulin-like growth factor binding protein 3 |
| chr10_-_28623368 | 0.08 |
ENST00000441595.2
|
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr3_-_194393206 | 0.08 |
ENST00000265245.5
|
LSG1
|
large 60S subunit nuclear export GTPase 1 |
| chr19_-_42947121 | 0.08 |
ENST00000601181.1
|
CXCL17
|
chemokine (C-X-C motif) ligand 17 |
| chr6_+_20534672 | 0.08 |
ENST00000274695.4
ENST00000378624.4 |
CDKAL1
|
CDK5 regulatory subunit associated protein 1-like 1 |
| chr5_+_146614579 | 0.07 |
ENST00000541094.1
ENST00000398521.3 |
STK32A
|
serine/threonine kinase 32A |
| chr3_+_130279178 | 0.07 |
ENST00000358511.6
ENST00000453409.2 |
COL6A6
|
collagen, type VI, alpha 6 |
| chr6_-_49712091 | 0.07 |
ENST00000371159.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr6_-_30585009 | 0.07 |
ENST00000376511.2
|
PPP1R10
|
protein phosphatase 1, regulatory subunit 10 |
| chr20_-_1309809 | 0.07 |
ENST00000360779.3
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr15_-_72523924 | 0.07 |
ENST00000566809.1
ENST00000567087.1 ENST00000569050.1 ENST00000568883.1 |
PKM
|
pyruvate kinase, muscle |
| chr15_+_93749295 | 0.07 |
ENST00000599897.1
|
AC112693.2
|
AC112693.2 |
| chr12_+_40787194 | 0.07 |
ENST00000425730.2
ENST00000454784.4 |
MUC19
|
mucin 19, oligomeric |
| chr19_+_30156351 | 0.07 |
ENST00000592810.1
|
PLEKHF1
|
pleckstrin homology domain containing, family F (with FYVE domain) member 1 |
| chr14_+_37641012 | 0.07 |
ENST00000556667.1
|
SLC25A21-AS1
|
SLC25A21 antisense RNA 1 |
| chr2_-_32489922 | 0.07 |
ENST00000402280.1
|
NLRC4
|
NLR family, CARD domain containing 4 |
| chrX_+_64808248 | 0.07 |
ENST00000609672.1
|
MSN
|
moesin |
| chr2_+_235887329 | 0.07 |
ENST00000409212.1
ENST00000344528.4 ENST00000444916.1 |
SH3BP4
|
SH3-domain binding protein 4 |
| chr1_-_85358850 | 0.07 |
ENST00000370611.3
|
LPAR3
|
lysophosphatidic acid receptor 3 |
| chr18_-_53019208 | 0.07 |
ENST00000562607.1
|
TCF4
|
transcription factor 4 |
| chrX_+_36254051 | 0.07 |
ENST00000378657.4
|
CXorf30
|
chromosome X open reading frame 30 |
| chr4_+_6271558 | 0.07 |
ENST00000503569.1
ENST00000226760.1 |
WFS1
|
Wolfram syndrome 1 (wolframin) |
| chr2_+_196313239 | 0.07 |
ENST00000413290.1
|
AC064834.1
|
AC064834.1 |
| chr11_-_26588634 | 0.07 |
ENST00000436318.2
ENST00000281268.8 |
MUC15
|
mucin 15, cell surface associated |
| chr4_+_71017791 | 0.07 |
ENST00000502294.1
|
C4orf40
|
chromosome 4 open reading frame 40 |
| chr17_+_40925454 | 0.07 |
ENST00000253794.2
ENST00000590339.1 ENST00000589520.1 |
VPS25
|
vacuolar protein sorting 25 homolog (S. cerevisiae) |
| chr10_+_134150835 | 0.07 |
ENST00000432555.2
|
LRRC27
|
leucine rich repeat containing 27 |
| chr16_-_29910365 | 0.07 |
ENST00000346932.5
ENST00000350527.3 ENST00000537485.1 ENST00000568380.1 |
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr2_-_69180012 | 0.07 |
ENST00000481498.1
|
GKN2
|
gastrokine 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ARID5A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.4 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 0.2 | GO:1900276 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.1 | 0.3 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.2 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.2 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.2 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.1 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.0 | 0.2 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.1 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.0 | 0.1 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.0 | 0.1 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.5 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.2 | GO:0048625 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) myoblast fate commitment(GO:0048625) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.2 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.2 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.2 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.3 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.2 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.0 | 0.1 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.1 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.0 | 0.1 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.0 | 0.1 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.0 | 0.4 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.2 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.0 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 | 0.0 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.1 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 0.1 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.2 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.2 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.1 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.0 | GO:0021816 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 0.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.2 | GO:0039713 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.2 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.1 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.2 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.2 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.0 | 0.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.2 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.1 | 0.4 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.2 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.1 | 0.3 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.0 | 0.2 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.1 | GO:0005018 | platelet-derived growth factor alpha-receptor activity(GO:0005018) |
| 0.0 | 0.1 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.2 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.7 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.0 | 0.2 | GO:0015562 | urate transmembrane transporter activity(GO:0015143) efflux transmembrane transporter activity(GO:0015562) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0004397 | histidine ammonia-lyase activity(GO:0004397) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.1 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.3 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.2 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 1.8 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |