Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
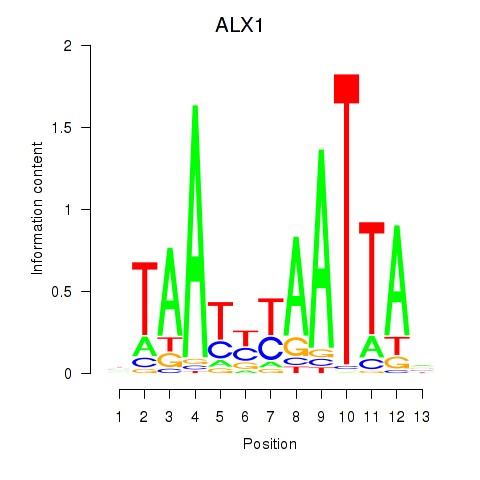
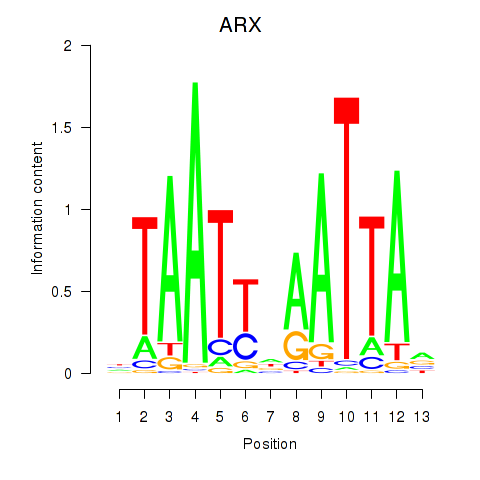
Results for ALX1_ARX
Z-value: 0.36
Transcription factors associated with ALX1_ARX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ALX1
|
ENSG00000180318.3 | ALX homeobox 1 |
|
ARX
|
ENSG00000004848.6 | aristaless related homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARX | hg19_v2_chrX_-_25034065_25034088 | 0.10 | 8.6e-01 | Click! |
| ALX1 | hg19_v2_chr12_+_85673868_85673885 | 0.07 | 9.0e-01 | Click! |
Activity profile of ALX1_ARX motif
Sorted Z-values of ALX1_ARX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_61151306 | 0.93 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr8_+_86999516 | 0.87 |
ENST00000521564.1
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr12_-_118796910 | 0.32 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr17_-_39023462 | 0.26 |
ENST00000251643.4
|
KRT12
|
keratin 12 |
| chr6_+_12007897 | 0.21 |
ENST00000437559.1
|
RP11-456H18.2
|
RP11-456H18.2 |
| chr14_-_95236551 | 0.21 |
ENST00000238558.3
|
GSC
|
goosecoid homeobox |
| chr7_-_111032971 | 0.21 |
ENST00000450877.1
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr1_-_234667504 | 0.20 |
ENST00000421207.1
ENST00000435574.1 |
RP5-855F14.1
|
RP5-855F14.1 |
| chr6_+_12007963 | 0.19 |
ENST00000607445.1
|
RP11-456H18.2
|
RP11-456H18.2 |
| chr14_-_57197224 | 0.19 |
ENST00000554597.1
ENST00000556696.1 |
RP11-1085N6.3
|
Uncharacterized protein |
| chr7_+_30589829 | 0.18 |
ENST00000579437.1
|
RP4-777O23.1
|
RP4-777O23.1 |
| chr12_-_91546926 | 0.18 |
ENST00000550758.1
|
DCN
|
decorin |
| chr6_-_26199499 | 0.18 |
ENST00000377831.5
|
HIST1H3D
|
histone cluster 1, H3d |
| chr3_-_157824292 | 0.15 |
ENST00000483851.2
|
SHOX2
|
short stature homeobox 2 |
| chr3_-_12587055 | 0.14 |
ENST00000564146.3
|
C3orf83
|
chromosome 3 open reading frame 83 |
| chr1_+_50459990 | 0.13 |
ENST00000448346.1
|
AL645730.2
|
AL645730.2 |
| chr12_-_118628315 | 0.13 |
ENST00000540561.1
|
TAOK3
|
TAO kinase 3 |
| chr2_+_29001711 | 0.13 |
ENST00000418910.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr11_-_82708435 | 0.13 |
ENST00000525117.1
ENST00000532548.1 |
RAB30
|
RAB30, member RAS oncogene family |
| chr10_+_97709725 | 0.11 |
ENST00000472454.2
|
RP11-248J23.6
|
Protein LOC100652732 |
| chr2_-_114461655 | 0.11 |
ENST00000424612.1
|
AC017074.2
|
AC017074.2 |
| chr12_+_14369524 | 0.11 |
ENST00000538329.1
|
RP11-134N1.2
|
RP11-134N1.2 |
| chr2_-_145188137 | 0.11 |
ENST00000440875.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr6_-_137314371 | 0.10 |
ENST00000432330.1
ENST00000418699.1 |
RP11-55K22.5
|
RP11-55K22.5 |
| chr1_-_151148442 | 0.10 |
ENST00000441701.1
ENST00000416280.2 |
TMOD4
|
tropomodulin 4 (muscle) |
| chr6_+_3259122 | 0.10 |
ENST00000438998.2
ENST00000380305.4 |
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4 |
| chr13_-_81801115 | 0.09 |
ENST00000567258.1
|
LINC00564
|
long intergenic non-protein coding RNA 564 |
| chr2_-_145275228 | 0.09 |
ENST00000427902.1
ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr12_+_116985896 | 0.08 |
ENST00000547114.1
|
RP11-809C9.2
|
RP11-809C9.2 |
| chr10_-_75226166 | 0.08 |
ENST00000544628.1
|
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr2_-_145275828 | 0.08 |
ENST00000392861.2
ENST00000409211.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr10_+_67330096 | 0.08 |
ENST00000433152.4
ENST00000601979.1 ENST00000599409.1 ENST00000608678.1 |
RP11-222A11.1
|
RP11-222A11.1 |
| chr18_+_616711 | 0.08 |
ENST00000579494.1
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr11_-_96076334 | 0.08 |
ENST00000524717.1
|
MAML2
|
mastermind-like 2 (Drosophila) |
| chr15_+_101417919 | 0.08 |
ENST00000561338.1
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3 |
| chr8_-_102803163 | 0.08 |
ENST00000523645.1
ENST00000520346.1 ENST00000220931.6 ENST00000522448.1 ENST00000522951.1 ENST00000522252.1 ENST00000519098.1 |
NCALD
|
neurocalcin delta |
| chr18_+_616672 | 0.07 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr8_-_57123815 | 0.07 |
ENST00000316981.3
ENST00000423799.2 ENST00000429357.2 |
PLAG1
|
pleiomorphic adenoma gene 1 |
| chr6_+_34204642 | 0.07 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr6_+_26087646 | 0.07 |
ENST00000309234.6
|
HFE
|
hemochromatosis |
| chr7_-_117512264 | 0.07 |
ENST00000454375.1
|
CTTNBP2
|
cortactin binding protein 2 |
| chr1_+_149239529 | 0.07 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chr1_+_15668240 | 0.07 |
ENST00000444385.1
|
FHAD1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr11_-_95523500 | 0.07 |
ENST00000540054.1
|
FAM76B
|
family with sequence similarity 76, member B |
| chr6_-_154751629 | 0.07 |
ENST00000424998.1
|
CNKSR3
|
CNKSR family member 3 |
| chr1_+_206557366 | 0.06 |
ENST00000414007.1
ENST00000419187.2 |
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr11_+_18433840 | 0.06 |
ENST00000541669.1
ENST00000280704.4 |
LDHC
|
lactate dehydrogenase C |
| chr6_-_138833630 | 0.06 |
ENST00000533765.1
|
NHSL1
|
NHS-like 1 |
| chr1_+_44679113 | 0.06 |
ENST00000361745.6
ENST00000446292.1 ENST00000440641.1 ENST00000436069.1 ENST00000437511.1 |
DMAP1
|
DNA methyltransferase 1 associated protein 1 |
| chr5_-_152069089 | 0.06 |
ENST00000506723.2
|
AC091969.1
|
AC091969.1 |
| chr1_+_244214577 | 0.06 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr4_+_118955500 | 0.06 |
ENST00000296499.5
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr13_+_74805561 | 0.06 |
ENST00000419499.1
|
LINC00402
|
long intergenic non-protein coding RNA 402 |
| chr17_+_71228843 | 0.06 |
ENST00000582391.1
|
C17orf80
|
chromosome 17 open reading frame 80 |
| chrX_+_107288239 | 0.06 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr12_-_49581152 | 0.06 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr5_-_87516448 | 0.05 |
ENST00000511218.1
|
TMEM161B
|
transmembrane protein 161B |
| chrX_+_107288197 | 0.05 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr1_+_44679159 | 0.05 |
ENST00000315913.5
ENST00000372289.2 |
DMAP1
|
DNA methyltransferase 1 associated protein 1 |
| chr7_-_55620433 | 0.05 |
ENST00000418904.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr1_+_145525015 | 0.05 |
ENST00000539363.1
ENST00000538811.1 |
ITGA10
|
integrin, alpha 10 |
| chr5_+_59783941 | 0.05 |
ENST00000506884.1
ENST00000504876.2 |
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr2_+_65215604 | 0.05 |
ENST00000531327.1
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr4_-_87028478 | 0.05 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr6_-_32908792 | 0.05 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr8_+_67039131 | 0.05 |
ENST00000315962.4
ENST00000353317.5 |
TRIM55
|
tripartite motif containing 55 |
| chr12_+_12878829 | 0.05 |
ENST00000326765.6
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr1_+_154401791 | 0.05 |
ENST00000476006.1
|
IL6R
|
interleukin 6 receptor |
| chr17_+_74463650 | 0.05 |
ENST00000392492.3
|
AANAT
|
aralkylamine N-acetyltransferase |
| chr19_-_6393465 | 0.04 |
ENST00000394456.5
|
GTF2F1
|
general transcription factor IIF, polypeptide 1, 74kDa |
| chr4_+_88571429 | 0.04 |
ENST00000339673.6
ENST00000282479.7 |
DMP1
|
dentin matrix acidic phosphoprotein 1 |
| chr6_-_133055896 | 0.04 |
ENST00000367927.5
ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3
|
vanin 3 |
| chr12_+_122688090 | 0.04 |
ENST00000324189.4
ENST00000546192.1 |
B3GNT4
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr18_+_20714525 | 0.04 |
ENST00000400473.2
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1 |
| chr8_+_107738343 | 0.04 |
ENST00000521592.1
|
OXR1
|
oxidation resistance 1 |
| chr22_-_30642728 | 0.04 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr5_-_111312622 | 0.04 |
ENST00000395634.3
|
NREP
|
neuronal regeneration related protein |
| chr4_+_76649753 | 0.04 |
ENST00000603759.1
|
USO1
|
USO1 vesicle transport factor |
| chr15_+_80733570 | 0.04 |
ENST00000533983.1
ENST00000527771.1 ENST00000525103.1 |
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr18_-_52989217 | 0.04 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr6_+_3259148 | 0.04 |
ENST00000419065.2
ENST00000473000.2 ENST00000451246.2 ENST00000454610.2 |
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4 |
| chr12_-_118796971 | 0.04 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3 |
| chr4_-_89951028 | 0.04 |
ENST00000506913.1
|
FAM13A
|
family with sequence similarity 13, member A |
| chr15_+_51669513 | 0.04 |
ENST00000558426.1
|
GLDN
|
gliomedin |
| chr3_-_47934234 | 0.04 |
ENST00000420772.2
|
MAP4
|
microtubule-associated protein 4 |
| chr5_+_140227048 | 0.04 |
ENST00000532602.1
|
PCDHA9
|
protocadherin alpha 9 |
| chr2_+_74685413 | 0.04 |
ENST00000233615.2
|
WBP1
|
WW domain binding protein 1 |
| chr6_+_88106840 | 0.04 |
ENST00000369570.4
|
C6orf164
|
chromosome 6 open reading frame 164 |
| chr2_+_196313239 | 0.04 |
ENST00000413290.1
|
AC064834.1
|
AC064834.1 |
| chr10_+_97803151 | 0.03 |
ENST00000403870.3
ENST00000265992.5 ENST00000465148.2 ENST00000534974.1 |
CCNJ
|
cyclin J |
| chr5_+_133562095 | 0.03 |
ENST00000602919.1
|
CTD-2410N18.3
|
CTD-2410N18.3 |
| chrX_+_36254051 | 0.03 |
ENST00000378657.4
|
CXorf30
|
chromosome X open reading frame 30 |
| chr6_-_133055815 | 0.03 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr11_+_1295809 | 0.03 |
ENST00000598274.1
|
AC136297.1
|
Uncharacterized protein |
| chr8_+_107738240 | 0.03 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr19_-_6393216 | 0.03 |
ENST00000595047.1
|
GTF2F1
|
general transcription factor IIF, polypeptide 1, 74kDa |
| chrX_-_48858667 | 0.03 |
ENST00000376423.4
ENST00000376441.1 |
GRIPAP1
|
GRIP1 associated protein 1 |
| chr3_-_45838011 | 0.03 |
ENST00000358525.4
ENST00000413781.1 |
SLC6A20
|
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr3_+_111393501 | 0.03 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr3_-_141747459 | 0.03 |
ENST00000477292.1
ENST00000478006.1 ENST00000495310.1 ENST00000486111.1 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr2_-_145277882 | 0.03 |
ENST00000465070.1
ENST00000444559.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr4_-_69083720 | 0.03 |
ENST00000432593.3
|
TMPRSS11BNL
|
TMPRSS11B N-terminal like |
| chr19_-_38183201 | 0.03 |
ENST00000590008.1
ENST00000358582.4 |
ZNF781
|
zinc finger protein 781 |
| chrX_-_48858630 | 0.03 |
ENST00000376425.3
ENST00000376444.3 |
GRIPAP1
|
GRIP1 associated protein 1 |
| chr5_+_59783540 | 0.03 |
ENST00000515734.2
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr19_-_54619006 | 0.03 |
ENST00000391759.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr17_+_7155819 | 0.03 |
ENST00000570322.1
ENST00000576496.1 ENST00000574841.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr11_-_95522907 | 0.02 |
ENST00000358780.5
ENST00000542135.1 |
FAM76B
|
family with sequence similarity 76, member B |
| chr10_-_29923893 | 0.02 |
ENST00000355867.4
|
SVIL
|
supervillin |
| chr1_-_232651312 | 0.02 |
ENST00000262861.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr17_-_39623681 | 0.02 |
ENST00000225899.3
|
KRT32
|
keratin 32 |
| chr1_+_170632250 | 0.02 |
ENST00000367760.3
|
PRRX1
|
paired related homeobox 1 |
| chr12_+_1099675 | 0.02 |
ENST00000545318.2
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr14_-_81425828 | 0.02 |
ENST00000555529.1
ENST00000556042.1 ENST00000556981.1 |
CEP128
|
centrosomal protein 128kDa |
| chr1_-_94147385 | 0.02 |
ENST00000260502.6
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr6_-_26199471 | 0.02 |
ENST00000341023.1
|
HIST1H2AD
|
histone cluster 1, H2ad |
| chr6_+_15401075 | 0.02 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr3_-_45837959 | 0.02 |
ENST00000353278.4
ENST00000456124.2 |
SLC6A20
|
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr15_-_75748143 | 0.02 |
ENST00000568431.1
ENST00000568309.1 ENST00000568190.1 ENST00000570115.1 ENST00000564778.1 |
SIN3A
|
SIN3 transcription regulator family member A |
| chr3_-_87325612 | 0.02 |
ENST00000561167.1
ENST00000560656.1 ENST00000344265.3 |
POU1F1
|
POU class 1 homeobox 1 |
| chr13_-_47471155 | 0.02 |
ENST00000543956.1
ENST00000542664.1 |
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
| chr11_-_102709441 | 0.02 |
ENST00000434103.1
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr3_-_101039402 | 0.02 |
ENST00000193391.7
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2 |
| chr3_-_27763803 | 0.02 |
ENST00000449599.1
|
EOMES
|
eomesodermin |
| chr8_-_6420930 | 0.02 |
ENST00000325203.5
|
ANGPT2
|
angiopoietin 2 |
| chr1_-_110155671 | 0.02 |
ENST00000351050.3
|
GNAT2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 |
| chr14_-_101036119 | 0.02 |
ENST00000355173.2
|
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr2_+_176994408 | 0.02 |
ENST00000429017.1
ENST00000313173.4 ENST00000544999.1 |
HOXD8
|
homeobox D8 |
| chr1_+_104615595 | 0.02 |
ENST00000418362.1
|
RP11-364B6.1
|
RP11-364B6.1 |
| chr5_+_137722255 | 0.02 |
ENST00000542866.1
|
KDM3B
|
lysine (K)-specific demethylase 3B |
| chr3_+_190105909 | 0.02 |
ENST00000456423.1
|
CLDN16
|
claudin 16 |
| chr8_-_40200903 | 0.02 |
ENST00000522486.1
|
CTA-392C11.1
|
CTA-392C11.1 |
| chr15_-_52970820 | 0.02 |
ENST00000261844.7
ENST00000399202.4 ENST00000562135.1 |
FAM214A
|
family with sequence similarity 214, member A |
| chr21_-_43346790 | 0.02 |
ENST00000329623.7
|
C2CD2
|
C2 calcium-dependent domain containing 2 |
| chr10_+_89420706 | 0.02 |
ENST00000427144.2
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr16_+_1728257 | 0.02 |
ENST00000248098.3
ENST00000562684.1 ENST00000561516.1 ENST00000382711.5 ENST00000566742.1 |
HN1L
|
hematological and neurological expressed 1-like |
| chr7_-_14880892 | 0.02 |
ENST00000406247.3
ENST00000399322.3 ENST00000258767.5 |
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr3_-_164914640 | 0.02 |
ENST00000241274.3
|
SLITRK3
|
SLIT and NTRK-like family, member 3 |
| chr11_+_85359062 | 0.02 |
ENST00000532180.1
|
TMEM126A
|
transmembrane protein 126A |
| chr6_-_170893268 | 0.02 |
ENST00000538195.1
|
PDCD2
|
programmed cell death 2 |
| chr7_+_90338712 | 0.02 |
ENST00000265741.3
ENST00000406263.1 |
CDK14
|
cyclin-dependent kinase 14 |
| chr5_+_140514782 | 0.01 |
ENST00000231134.5
|
PCDHB5
|
protocadherin beta 5 |
| chr5_-_160279207 | 0.01 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr9_-_27005686 | 0.01 |
ENST00000380055.5
|
LRRC19
|
leucine rich repeat containing 19 |
| chr7_-_115799942 | 0.01 |
ENST00000484212.1
|
TFEC
|
transcription factor EC |
| chr3_+_39424828 | 0.01 |
ENST00000273158.4
ENST00000431510.1 |
SLC25A38
|
solute carrier family 25, member 38 |
| chr17_+_71228537 | 0.01 |
ENST00000577615.1
ENST00000585109.1 |
C17orf80
|
chromosome 17 open reading frame 80 |
| chr2_-_145278475 | 0.01 |
ENST00000558170.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr5_-_55412774 | 0.01 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr14_+_24099318 | 0.01 |
ENST00000432832.2
|
DHRS2
|
dehydrogenase/reductase (SDR family) member 2 |
| chr7_-_7575477 | 0.01 |
ENST00000399429.3
|
COL28A1
|
collagen, type XXVIII, alpha 1 |
| chr12_+_59989918 | 0.01 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chrX_+_39868501 | 0.01 |
ENST00000447651.1
|
AC092198.1
|
AC092198.1 |
| chr2_+_234600253 | 0.01 |
ENST00000373424.1
ENST00000441351.1 |
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr15_+_42066632 | 0.01 |
ENST00000457542.2
ENST00000221214.6 ENST00000260357.7 ENST00000456763.2 |
MAPKBP1
|
mitogen-activated protein kinase binding protein 1 |
| chr11_+_85359002 | 0.01 |
ENST00000528105.1
ENST00000304511.2 |
TMEM126A
|
transmembrane protein 126A |
| chr11_+_95523621 | 0.01 |
ENST00000325542.5
ENST00000325486.5 ENST00000544522.1 ENST00000541365.1 |
CEP57
|
centrosomal protein 57kDa |
| chr6_+_31895254 | 0.01 |
ENST00000299367.5
ENST00000442278.2 |
C2
|
complement component 2 |
| chr9_-_95298254 | 0.01 |
ENST00000444490.2
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr2_+_228678550 | 0.01 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr14_+_38264418 | 0.01 |
ENST00000267368.7
ENST00000382320.3 |
TTC6
|
tetratricopeptide repeat domain 6 |
| chr4_-_122854612 | 0.01 |
ENST00000264811.5
|
TRPC3
|
transient receptor potential cation channel, subfamily C, member 3 |
| chr7_+_136553370 | 0.01 |
ENST00000445907.2
|
CHRM2
|
cholinergic receptor, muscarinic 2 |
| chr2_+_101437487 | 0.01 |
ENST00000427413.1
ENST00000542504.1 |
NPAS2
|
neuronal PAS domain protein 2 |
| chr10_+_134150835 | 0.01 |
ENST00000432555.2
|
LRRC27
|
leucine rich repeat containing 27 |
| chr2_-_31440377 | 0.01 |
ENST00000444918.2
ENST00000403897.3 |
CAPN14
|
calpain 14 |
| chr8_-_91095099 | 0.01 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr13_-_45775162 | 0.01 |
ENST00000405872.1
|
KCTD4
|
potassium channel tetramerization domain containing 4 |
| chr11_+_120973375 | 0.01 |
ENST00000264037.2
|
TECTA
|
tectorin alpha |
| chr18_-_13915530 | 0.01 |
ENST00000327606.3
|
MC2R
|
melanocortin 2 receptor (adrenocorticotropic hormone) |
| chr18_-_52989525 | 0.01 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr19_+_50016610 | 0.01 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chrX_+_36246735 | 0.01 |
ENST00000378653.3
|
CXorf30
|
chromosome X open reading frame 30 |
| chr12_-_89746264 | 0.01 |
ENST00000548755.1
|
DUSP6
|
dual specificity phosphatase 6 |
| chr2_+_162272605 | 0.01 |
ENST00000389554.3
|
TBR1
|
T-box, brain, 1 |
| chr15_+_51669444 | 0.01 |
ENST00000396399.2
|
GLDN
|
gliomedin |
| chr8_+_39792474 | 0.01 |
ENST00000502986.2
|
IDO2
|
indoleamine 2,3-dioxygenase 2 |
| chr15_+_44092784 | 0.01 |
ENST00000458412.1
|
HYPK
|
huntingtin interacting protein K |
| chr17_-_39123144 | 0.01 |
ENST00000355612.2
|
KRT39
|
keratin 39 |
| chr3_-_124774802 | 0.01 |
ENST00000311127.4
|
HEG1
|
heart development protein with EGF-like domains 1 |
| chr15_+_44719790 | 0.01 |
ENST00000558791.1
|
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr21_+_17442799 | 0.01 |
ENST00000602580.1
ENST00000458468.1 ENST00000602935.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr9_+_44867571 | 0.01 |
ENST00000377548.2
|
RP11-160N1.10
|
RP11-160N1.10 |
| chr9_+_88556444 | 0.01 |
ENST00000376040.1
|
NAA35
|
N(alpha)-acetyltransferase 35, NatC auxiliary subunit |
| chr7_-_33842742 | 0.01 |
ENST00000420185.1
ENST00000440034.1 |
RP11-89N17.4
|
RP11-89N17.4 |
| chr3_+_140981456 | 0.01 |
ENST00000504264.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr2_+_54683419 | 0.01 |
ENST00000356805.4
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr10_+_53806501 | 0.01 |
ENST00000373975.2
|
PRKG1
|
protein kinase, cGMP-dependent, type I |
| chr6_-_107235378 | 0.01 |
ENST00000606430.1
|
RP1-60O19.1
|
RP1-60O19.1 |
| chr12_+_122241928 | 0.01 |
ENST00000604567.1
ENST00000542440.1 |
SETD1B
|
SET domain containing 1B |
| chr17_-_71228357 | 0.00 |
ENST00000583024.1
ENST00000403627.3 ENST00000405159.3 ENST00000581110.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr11_+_128563652 | 0.00 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr9_-_71869642 | 0.00 |
ENST00000600472.1
|
AL358113.1
|
Tight junction protein 2 (Zona occludens 2) isoform 1 variant; Uncharacterized protein |
| chr7_-_82792215 | 0.00 |
ENST00000333891.9
ENST00000423517.2 |
PCLO
|
piccolo presynaptic cytomatrix protein |
| chr5_-_115872142 | 0.00 |
ENST00000510263.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr5_+_140762268 | 0.00 |
ENST00000518325.1
|
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chrX_-_138724994 | 0.00 |
ENST00000536274.1
|
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr7_+_90339169 | 0.00 |
ENST00000436577.2
|
CDK14
|
cyclin-dependent kinase 14 |
| chr6_+_53964336 | 0.00 |
ENST00000447836.2
ENST00000511678.1 |
MLIP
|
muscular LMNA-interacting protein |
| chr11_+_22694123 | 0.00 |
ENST00000534801.1
|
GAS2
|
growth arrest-specific 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ALX1_ARX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.3 | GO:1902748 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.1 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.9 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.1 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.0 | 0.1 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.0 | 0.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:0021816 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.0 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.1 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 0.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.4 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.0 | GO:0002384 | hepatic immune response(GO:0002384) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.9 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.0 | 0.0 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.0 | 0.0 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.0 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |