Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
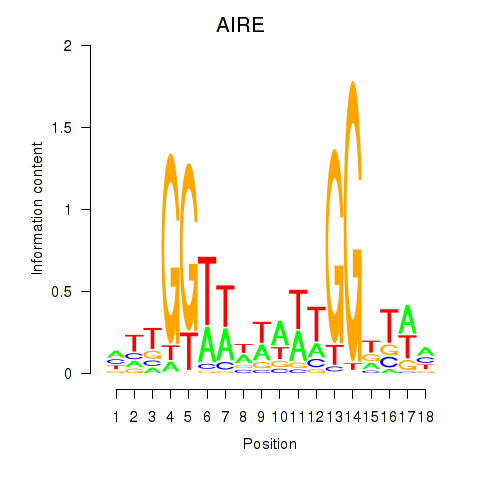
Results for AIRE
Z-value: 0.54
Transcription factors associated with AIRE
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
AIRE
|
ENSG00000160224.12 | autoimmune regulator |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| AIRE | hg19_v2_chr21_+_45705752_45705767 | -0.62 | 1.9e-01 | Click! |
Activity profile of AIRE motif
Sorted Z-values of AIRE motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrM_+_8366 | 0.59 |
ENST00000361851.1
|
MT-ATP8
|
mitochondrially encoded ATP synthase 8 |
| chr1_+_79115503 | 0.57 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr2_+_38177575 | 0.53 |
ENST00000407257.1
ENST00000417700.2 ENST00000234195.3 ENST00000442857.1 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chr16_+_56672571 | 0.45 |
ENST00000290705.8
|
MT1A
|
metallothionein 1A |
| chr11_+_45376922 | 0.44 |
ENST00000524410.1
ENST00000524488.1 ENST00000524565.1 |
RP11-430H10.1
|
RP11-430H10.1 |
| chr7_-_64023441 | 0.42 |
ENST00000309683.6
|
ZNF680
|
zinc finger protein 680 |
| chr6_+_26104104 | 0.41 |
ENST00000377803.2
|
HIST1H4C
|
histone cluster 1, H4c |
| chr17_-_39150385 | 0.37 |
ENST00000391586.1
|
KRTAP3-3
|
keratin associated protein 3-3 |
| chr5_-_38845812 | 0.36 |
ENST00000513480.1
ENST00000512519.1 |
CTD-2127H9.1
|
CTD-2127H9.1 |
| chrM_+_7586 | 0.33 |
ENST00000361739.1
|
MT-CO2
|
mitochondrially encoded cytochrome c oxidase II |
| chr4_-_104119528 | 0.31 |
ENST00000380026.3
ENST00000503705.1 ENST00000265148.3 |
CENPE
|
centromere protein E, 312kDa |
| chrM_+_14741 | 0.30 |
ENST00000361789.2
|
MT-CYB
|
mitochondrially encoded cytochrome b |
| chr16_+_50059182 | 0.29 |
ENST00000562576.1
|
CNEP1R1
|
CTD nuclear envelope phosphatase 1 regulatory subunit 1 |
| chr22_+_36649170 | 0.28 |
ENST00000438034.1
ENST00000427990.1 ENST00000347595.7 ENST00000397279.4 ENST00000433768.1 ENST00000440669.2 |
APOL1
|
apolipoprotein L, 1 |
| chr14_-_55658323 | 0.27 |
ENST00000554067.1
ENST00000247191.2 |
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr6_-_127663543 | 0.25 |
ENST00000531582.1
|
ECHDC1
|
enoyl CoA hydratase domain containing 1 |
| chr22_+_25202232 | 0.24 |
ENST00000400358.4
ENST00000400359.4 |
SGSM1
|
small G protein signaling modulator 1 |
| chr14_-_55658252 | 0.23 |
ENST00000395425.2
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr19_-_52551814 | 0.23 |
ENST00000594154.1
ENST00000598745.1 ENST00000597273.1 |
ZNF432
|
zinc finger protein 432 |
| chr7_+_116451100 | 0.23 |
ENST00000464223.1
ENST00000484325.1 |
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr12_-_76462713 | 0.22 |
ENST00000552056.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr6_+_168434678 | 0.22 |
ENST00000496008.1
|
KIF25
|
kinesin family member 25 |
| chr11_-_8954491 | 0.22 |
ENST00000526227.1
ENST00000525780.1 ENST00000326053.5 |
C11orf16
|
chromosome 11 open reading frame 16 |
| chr6_-_31514516 | 0.22 |
ENST00000303892.5
ENST00000483251.1 |
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr10_-_4720333 | 0.21 |
ENST00000430998.2
|
LINC00704
|
long intergenic non-protein coding RNA 704 |
| chr7_+_142374104 | 0.21 |
ENST00000604952.1
|
MTRNR2L6
|
MT-RNR2-like 6 |
| chr1_-_154600421 | 0.20 |
ENST00000368471.3
ENST00000292205.5 |
ADAR
|
adenosine deaminase, RNA-specific |
| chr12_+_20963647 | 0.20 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr19_-_23869970 | 0.20 |
ENST00000601010.1
|
ZNF675
|
zinc finger protein 675 |
| chr11_+_6947647 | 0.20 |
ENST00000278319.5
|
ZNF215
|
zinc finger protein 215 |
| chr4_-_47465666 | 0.20 |
ENST00000381571.4
|
COMMD8
|
COMM domain containing 8 |
| chr6_-_106773610 | 0.18 |
ENST00000369076.3
ENST00000369070.1 |
ATG5
|
autophagy related 5 |
| chr4_+_76649797 | 0.17 |
ENST00000538159.1
ENST00000514213.2 |
USO1
|
USO1 vesicle transport factor |
| chr14_+_45605157 | 0.17 |
ENST00000542564.2
|
FANCM
|
Fanconi anemia, complementation group M |
| chr5_+_118691008 | 0.17 |
ENST00000504642.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr14_+_45605127 | 0.17 |
ENST00000556036.1
ENST00000267430.5 |
FANCM
|
Fanconi anemia, complementation group M |
| chrM_+_12331 | 0.17 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chrM_-_14670 | 0.17 |
ENST00000361681.2
|
MT-ND6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chrM_+_5824 | 0.17 |
ENST00000361624.2
|
MT-CO1
|
mitochondrially encoded cytochrome c oxidase I |
| chr5_+_32788945 | 0.17 |
ENST00000326958.1
|
AC026703.1
|
AC026703.1 |
| chrM_+_4431 | 0.16 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr1_-_52344416 | 0.16 |
ENST00000544028.1
|
NRD1
|
nardilysin (N-arginine dibasic convertase) |
| chr16_+_75182376 | 0.16 |
ENST00000570010.1
ENST00000568079.1 ENST00000464850.1 ENST00000332307.4 ENST00000393430.2 |
ZFP1
|
ZFP1 zinc finger protein |
| chr16_-_31147020 | 0.16 |
ENST00000568261.1
ENST00000567797.1 ENST00000317508.6 |
PRSS8
|
protease, serine, 8 |
| chr8_+_104384616 | 0.15 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr12_+_75874460 | 0.15 |
ENST00000266659.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr6_-_31514333 | 0.15 |
ENST00000376151.4
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr3_-_19975665 | 0.14 |
ENST00000295824.9
ENST00000389256.4 |
EFHB
|
EF-hand domain family, member B |
| chr14_+_58894706 | 0.14 |
ENST00000261244.5
|
KIAA0586
|
KIAA0586 |
| chr2_-_166810261 | 0.14 |
ENST00000243344.7
|
TTC21B
|
tetratricopeptide repeat domain 21B |
| chr4_-_114682719 | 0.13 |
ENST00000394522.3
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta |
| chr19_-_16606988 | 0.13 |
ENST00000269881.3
|
CALR3
|
calreticulin 3 |
| chr5_+_72143988 | 0.13 |
ENST00000506351.2
|
TNPO1
|
transportin 1 |
| chr19_-_48753028 | 0.12 |
ENST00000522431.1
|
CARD8
|
caspase recruitment domain family, member 8 |
| chr11_+_94038759 | 0.12 |
ENST00000440961.2
ENST00000328458.4 |
FOLR4
|
folate receptor 4, delta (putative) |
| chr4_-_4544061 | 0.12 |
ENST00000507908.1
|
STX18
|
syntaxin 18 |
| chr1_-_161208013 | 0.12 |
ENST00000515452.1
ENST00000367983.4 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr1_+_198189921 | 0.12 |
ENST00000391974.3
|
NEK7
|
NIMA-related kinase 7 |
| chr15_+_83209620 | 0.11 |
ENST00000568285.1
|
RP11-379H8.1
|
Uncharacterized protein |
| chr15_+_52043758 | 0.11 |
ENST00000249700.4
ENST00000539962.2 |
TMOD2
|
tropomodulin 2 (neuronal) |
| chr8_+_124780672 | 0.11 |
ENST00000521166.1
ENST00000334705.7 |
FAM91A1
|
family with sequence similarity 91, member A1 |
| chr10_-_106240032 | 0.11 |
ENST00000447860.1
|
RP11-127O4.3
|
RP11-127O4.3 |
| chr5_+_118690466 | 0.11 |
ENST00000503646.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr8_+_86851932 | 0.11 |
ENST00000517368.1
|
CTA-392E5.1
|
CTA-392E5.1 |
| chr4_+_156775910 | 0.11 |
ENST00000506072.1
ENST00000507590.1 |
TDO2
|
tryptophan 2,3-dioxygenase |
| chr2_+_172864490 | 0.10 |
ENST00000315796.4
|
METAP1D
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr15_+_27112948 | 0.10 |
ENST00000555060.1
|
GABRA5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr12_+_10658489 | 0.10 |
ENST00000538173.1
|
EIF2S3L
|
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr12_+_12202785 | 0.10 |
ENST00000586576.1
ENST00000464885.2 |
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr11_+_35211511 | 0.09 |
ENST00000524922.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr21_+_30673091 | 0.09 |
ENST00000447177.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr15_+_52043813 | 0.09 |
ENST00000435126.2
|
TMOD2
|
tropomodulin 2 (neuronal) |
| chr3_+_88199099 | 0.09 |
ENST00000486971.1
|
C3orf38
|
chromosome 3 open reading frame 38 |
| chr11_-_119249805 | 0.09 |
ENST00000527843.1
|
USP2
|
ubiquitin specific peptidase 2 |
| chr7_-_15601595 | 0.08 |
ENST00000342526.3
|
AGMO
|
alkylglycerol monooxygenase |
| chr20_+_12989596 | 0.08 |
ENST00000434210.1
ENST00000399002.2 |
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr4_+_66536248 | 0.08 |
ENST00000514260.1
ENST00000507117.1 |
RP11-807H7.1
|
RP11-807H7.1 |
| chr9_-_4666421 | 0.08 |
ENST00000381895.5
|
SPATA6L
|
spermatogenesis associated 6-like |
| chr20_+_21284416 | 0.08 |
ENST00000539513.1
|
XRN2
|
5'-3' exoribonuclease 2 |
| chr2_-_202508169 | 0.08 |
ENST00000409883.2
|
TMEM237
|
transmembrane protein 237 |
| chr2_-_127977654 | 0.08 |
ENST00000409327.1
|
CYP27C1
|
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr16_+_50059125 | 0.08 |
ENST00000427478.2
|
CNEP1R1
|
CTD nuclear envelope phosphatase 1 regulatory subunit 1 |
| chr1_+_100810575 | 0.08 |
ENST00000542213.1
|
CDC14A
|
cell division cycle 14A |
| chr18_-_37380230 | 0.08 |
ENST00000591629.1
|
LINC00669
|
long intergenic non-protein coding RNA 669 |
| chr15_-_52043631 | 0.07 |
ENST00000558126.1
|
LYSMD2
|
LysM, putative peptidoglycan-binding, domain containing 2 |
| chr1_-_145826450 | 0.07 |
ENST00000462900.2
|
GPR89A
|
G protein-coupled receptor 89A |
| chr19_+_45417921 | 0.07 |
ENST00000252491.4
ENST00000592885.1 ENST00000589781.1 |
APOC1
|
apolipoprotein C-I |
| chr6_+_26538566 | 0.07 |
ENST00000377575.2
|
HMGN4
|
high mobility group nucleosomal binding domain 4 |
| chr19_-_36643329 | 0.07 |
ENST00000589154.1
|
COX7A1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr22_+_36649056 | 0.07 |
ENST00000397278.3
ENST00000422706.1 ENST00000426053.1 ENST00000319136.4 |
APOL1
|
apolipoprotein L, 1 |
| chr20_+_44036620 | 0.07 |
ENST00000372710.3
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr6_-_169582835 | 0.07 |
ENST00000564830.1
|
RP11-417E7.2
|
RP11-417E7.2 |
| chr5_+_122181184 | 0.07 |
ENST00000513881.1
|
SNX24
|
sorting nexin 24 |
| chr9_-_114361919 | 0.06 |
ENST00000422125.1
|
PTGR1
|
prostaglandin reductase 1 |
| chr13_-_46756351 | 0.06 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr16_+_50914085 | 0.06 |
ENST00000576842.1
|
CTD-2034I21.2
|
CTD-2034I21.2 |
| chr19_-_21950362 | 0.06 |
ENST00000358296.6
|
ZNF100
|
zinc finger protein 100 |
| chr9_-_114362093 | 0.06 |
ENST00000538962.1
ENST00000407693.2 ENST00000238248.3 |
PTGR1
|
prostaglandin reductase 1 |
| chr3_-_123710199 | 0.06 |
ENST00000184183.4
|
ROPN1
|
rhophilin associated tail protein 1 |
| chr9_-_114361665 | 0.06 |
ENST00000309195.5
|
PTGR1
|
prostaglandin reductase 1 |
| chr11_-_56310757 | 0.06 |
ENST00000528616.2
|
OR5M11
|
olfactory receptor, family 5, subfamily M, member 11 |
| chr16_+_3254247 | 0.05 |
ENST00000304646.2
|
OR1F1
|
olfactory receptor, family 1, subfamily F, member 1 |
| chr4_-_76649546 | 0.05 |
ENST00000508510.1
ENST00000509561.1 ENST00000499709.2 ENST00000511868.1 |
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr4_+_78829479 | 0.05 |
ENST00000504901.1
|
MRPL1
|
mitochondrial ribosomal protein L1 |
| chr5_+_140625147 | 0.05 |
ENST00000231173.3
|
PCDHB15
|
protocadherin beta 15 |
| chr12_-_11287243 | 0.05 |
ENST00000539585.1
|
TAS2R30
|
taste receptor, type 2, member 30 |
| chr1_-_161207986 | 0.05 |
ENST00000506209.1
ENST00000367980.2 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr2_+_62423242 | 0.05 |
ENST00000301998.4
|
B3GNT2
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 |
| chr16_-_31146961 | 0.05 |
ENST00000567531.1
|
PRSS8
|
protease, serine, 8 |
| chr4_-_120222076 | 0.05 |
ENST00000504110.1
|
C4orf3
|
chromosome 4 open reading frame 3 |
| chr2_-_89568263 | 0.05 |
ENST00000473726.1
|
IGKV1-33
|
immunoglobulin kappa variable 1-33 |
| chr3_+_107318157 | 0.05 |
ENST00000406780.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chr9_-_113761720 | 0.05 |
ENST00000541779.1
ENST00000374430.2 |
LPAR1
|
lysophosphatidic acid receptor 1 |
| chr5_-_36242119 | 0.04 |
ENST00000511088.1
ENST00000282512.3 ENST00000506945.1 |
NADK2
|
NAD kinase 2, mitochondrial |
| chr12_+_7282795 | 0.04 |
ENST00000266546.6
|
CLSTN3
|
calsyntenin 3 |
| chr5_+_122181128 | 0.04 |
ENST00000261369.4
|
SNX24
|
sorting nexin 24 |
| chrX_-_102941596 | 0.04 |
ENST00000441076.2
ENST00000422355.1 ENST00000442614.1 ENST00000422154.2 ENST00000451301.1 |
MORF4L2
|
mortality factor 4 like 2 |
| chr17_+_34171081 | 0.04 |
ENST00000585577.1
|
TAF15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa |
| chr4_-_123843597 | 0.04 |
ENST00000510735.1
ENST00000304430.5 |
NUDT6
|
nudix (nucleoside diphosphate linked moiety X)-type motif 6 |
| chr2_+_179316163 | 0.04 |
ENST00000409117.3
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr17_+_77030267 | 0.04 |
ENST00000581774.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr2_+_1417228 | 0.04 |
ENST00000382269.3
ENST00000337415.3 ENST00000345913.4 ENST00000346956.3 ENST00000349624.3 ENST00000539820.1 ENST00000329066.4 ENST00000382201.3 |
TPO
|
thyroid peroxidase |
| chr12_+_12202774 | 0.04 |
ENST00000589718.1
|
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr1_-_52344471 | 0.04 |
ENST00000352171.7
ENST00000354831.7 |
NRD1
|
nardilysin (N-arginine dibasic convertase) |
| chr19_+_11455900 | 0.04 |
ENST00000588790.1
|
CCDC159
|
coiled-coil domain containing 159 |
| chr12_-_75603482 | 0.03 |
ENST00000341669.3
ENST00000298972.1 ENST00000350228.2 |
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr19_+_45417812 | 0.03 |
ENST00000592535.1
|
APOC1
|
apolipoprotein C-I |
| chr14_-_65409438 | 0.03 |
ENST00000557049.1
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr11_-_18956556 | 0.03 |
ENST00000302797.3
|
MRGPRX1
|
MAS-related GPR, member X1 |
| chr7_-_36764142 | 0.03 |
ENST00000258749.5
ENST00000535891.1 |
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr19_-_23869999 | 0.03 |
ENST00000601935.1
ENST00000359788.4 ENST00000600313.1 ENST00000596211.1 ENST00000599168.1 |
ZNF675
|
zinc finger protein 675 |
| chr21_+_39644214 | 0.03 |
ENST00000438657.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr4_+_71587669 | 0.03 |
ENST00000381006.3
ENST00000226328.4 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr7_-_87936195 | 0.03 |
ENST00000414498.1
ENST00000301959.5 ENST00000380079.4 |
STEAP4
|
STEAP family member 4 |
| chrX_-_153192211 | 0.03 |
ENST00000461052.1
ENST00000422091.1 |
ARHGAP4
|
Rho GTPase activating protein 4 |
| chr1_-_167883327 | 0.03 |
ENST00000476818.2
ENST00000367851.4 ENST00000367848.1 |
ADCY10
|
adenylate cyclase 10 (soluble) |
| chr16_-_71518504 | 0.03 |
ENST00000565100.2
|
ZNF19
|
zinc finger protein 19 |
| chr19_+_45418067 | 0.02 |
ENST00000589078.1
ENST00000586638.1 |
APOC1
|
apolipoprotein C-I |
| chr12_-_50677255 | 0.02 |
ENST00000551691.1
ENST00000394943.3 ENST00000341247.4 |
LIMA1
|
LIM domain and actin binding 1 |
| chr4_-_66536196 | 0.02 |
ENST00000511294.1
|
EPHA5
|
EPH receptor A5 |
| chr15_-_52043722 | 0.02 |
ENST00000454181.2
|
LYSMD2
|
LysM, putative peptidoglycan-binding, domain containing 2 |
| chr10_+_90660832 | 0.02 |
ENST00000371924.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr2_-_166060552 | 0.02 |
ENST00000283254.7
ENST00000453007.1 |
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr19_-_52552051 | 0.02 |
ENST00000221315.5
|
ZNF432
|
zinc finger protein 432 |
| chr14_-_106668095 | 0.02 |
ENST00000390606.2
|
IGHV3-20
|
immunoglobulin heavy variable 3-20 |
| chr12_+_48516463 | 0.02 |
ENST00000546465.1
|
PFKM
|
phosphofructokinase, muscle |
| chr9_-_21482312 | 0.02 |
ENST00000448696.3
|
IFNE
|
interferon, epsilon |
| chr3_-_121468602 | 0.02 |
ENST00000340645.5
|
GOLGB1
|
golgin B1 |
| chr7_+_20686946 | 0.02 |
ENST00000443026.2
ENST00000406935.1 |
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr12_-_64784471 | 0.01 |
ENST00000333722.5
|
C12orf56
|
chromosome 12 open reading frame 56 |
| chr4_-_16085657 | 0.01 |
ENST00000543373.1
|
PROM1
|
prominin 1 |
| chr21_+_30672433 | 0.01 |
ENST00000451655.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr6_-_33266687 | 0.01 |
ENST00000444031.2
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr19_-_16606930 | 0.01 |
ENST00000600762.1
|
CALR3
|
calreticulin 3 |
| chr19_+_56751773 | 0.01 |
ENST00000600684.1
|
ZSCAN5D
|
zinc finger and SCAN domain containing 5D |
| chr8_-_7220490 | 0.01 |
ENST00000400078.2
|
ZNF705G
|
zinc finger protein 705G |
| chr3_-_121264848 | 0.01 |
ENST00000264233.5
|
POLQ
|
polymerase (DNA directed), theta |
| chr4_-_57524061 | 0.01 |
ENST00000508121.1
|
HOPX
|
HOP homeobox |
| chr1_+_207669573 | 0.01 |
ENST00000400960.2
ENST00000534202.1 |
CR1
|
complement component (3b/4b) receptor 1 (Knops blood group) |
| chr12_+_20848486 | 0.01 |
ENST00000545102.1
|
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chrX_-_100307043 | 0.01 |
ENST00000372939.1
ENST00000372935.1 ENST00000372936.3 |
TRMT2B
|
tRNA methyltransferase 2 homolog B (S. cerevisiae) |
| chr21_+_39644395 | 0.01 |
ENST00000398934.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr8_+_54764346 | 0.01 |
ENST00000297313.3
ENST00000344277.6 |
RGS20
|
regulator of G-protein signaling 20 |
| chr3_-_33686925 | 0.01 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr4_-_120243545 | 0.00 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr21_+_39644172 | 0.00 |
ENST00000398932.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr16_+_72088376 | 0.00 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr5_-_107703556 | 0.00 |
ENST00000496714.1
|
FBXL17
|
F-box and leucine-rich repeat protein 17 |
| chr15_-_38519066 | 0.00 |
ENST00000561320.1
ENST00000561161.1 |
RP11-346D14.1
|
RP11-346D14.1 |
| chrX_-_153191708 | 0.00 |
ENST00000393721.1
ENST00000370028.3 |
ARHGAP4
|
Rho GTPase activating protein 4 |
| chr16_-_89119355 | 0.00 |
ENST00000537498.1
|
CTD-2555A7.2
|
CTD-2555A7.2 |
| chr19_-_48752812 | 0.00 |
ENST00000359009.4
|
CARD8
|
caspase recruitment domain family, member 8 |
| chr3_-_121468513 | 0.00 |
ENST00000494517.1
ENST00000393667.3 |
GOLGB1
|
golgin B1 |
| chr17_+_36452989 | 0.00 |
ENST00000312513.5
ENST00000582535.1 |
MRPL45
|
mitochondrial ribosomal protein L45 |
| chr9_+_90112117 | 0.00 |
ENST00000358077.5
|
DAPK1
|
death-associated protein kinase 1 |
| chr8_-_141728444 | 0.00 |
ENST00000521562.1
|
PTK2
|
protein tyrosine kinase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of AIRE
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.1 | 0.2 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.1 | 0.2 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.1 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.1 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.0 | 0.2 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.0 | 0.2 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.2 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.2 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.6 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.4 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.3 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.4 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 0.4 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.5 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.6 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.2 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.3 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.5 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 0.4 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.1 | 0.2 | GO:0032440 | 2-alkenal reductase [NAD(P)] activity(GO:0032440) |
| 0.0 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.3 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.2 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.1 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.6 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |