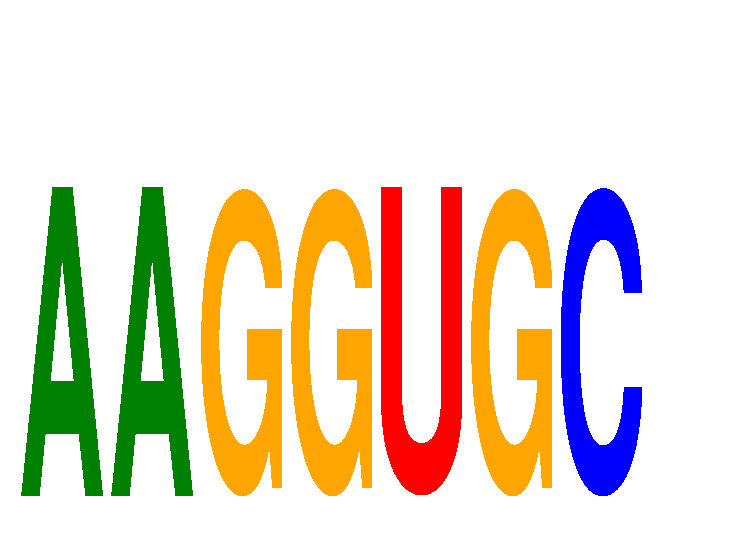Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for AAGGUGC
Z-value: 0.24
miRNA associated with seed AAGGUGC
| Name | miRBASE accession |
|---|---|
|
hsa-miR-18a-5p
|
MIMAT0000072 |
|
hsa-miR-18b-5p
|
MIMAT0001412 |
|
hsa-miR-4735-3p
|
MIMAT0019861 |
Activity profile of AAGGUGC motif
Sorted Z-values of AAGGUGC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_119806085 | 0.13 |
ENST00000355624.3
|
RAB11FIP2
|
RAB11 family interacting protein 2 (class I) |
| chr3_+_187930719 | 0.12 |
ENST00000312675.4
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr5_-_59189545 | 0.12 |
ENST00000340635.6
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr16_+_69599861 | 0.10 |
ENST00000354436.2
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr11_+_108093839 | 0.10 |
ENST00000452508.2
|
ATM
|
ataxia telangiectasia mutated |
| chrX_+_150565653 | 0.09 |
ENST00000330374.6
|
VMA21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr5_+_78532003 | 0.09 |
ENST00000396137.4
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr2_-_153032484 | 0.08 |
ENST00000263904.4
|
STAM2
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 |
| chr2_+_46926048 | 0.08 |
ENST00000306503.5
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr11_+_109964087 | 0.07 |
ENST00000278590.3
|
ZC3H12C
|
zinc finger CCCH-type containing 12C |
| chr3_+_107241783 | 0.07 |
ENST00000415149.2
ENST00000402543.1 ENST00000325805.8 ENST00000427402.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr15_-_56209306 | 0.07 |
ENST00000506154.1
ENST00000338963.2 ENST00000508342.1 |
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
| chr7_-_121036337 | 0.06 |
ENST00000426156.1
ENST00000359943.3 ENST00000412653.1 |
FAM3C
|
family with sequence similarity 3, member C |
| chr4_-_125633876 | 0.06 |
ENST00000504087.1
ENST00000515641.1 |
ANKRD50
|
ankyrin repeat domain 50 |
| chr11_-_130184555 | 0.06 |
ENST00000525842.1
|
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr2_-_206950781 | 0.06 |
ENST00000403263.1
|
INO80D
|
INO80 complex subunit D |
| chr5_-_43313574 | 0.06 |
ENST00000325110.6
ENST00000433297.2 |
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr5_-_180288248 | 0.06 |
ENST00000512132.1
ENST00000506439.1 ENST00000502412.1 ENST00000359141.6 |
ZFP62
|
ZFP62 zinc finger protein |
| chr10_+_111767720 | 0.06 |
ENST00000356080.4
ENST00000277900.8 |
ADD3
|
adducin 3 (gamma) |
| chr1_+_162467595 | 0.05 |
ENST00000538489.1
ENST00000489294.1 |
UHMK1
|
U2AF homology motif (UHM) kinase 1 |
| chr3_+_152017181 | 0.05 |
ENST00000498502.1
ENST00000324196.5 ENST00000545754.1 ENST00000357472.3 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr12_+_104458235 | 0.04 |
ENST00000229330.4
|
HCFC2
|
host cell factor C2 |
| chr3_-_113465065 | 0.04 |
ENST00000497255.1
ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr6_+_122720681 | 0.04 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chr20_+_49348081 | 0.04 |
ENST00000371610.2
|
PARD6B
|
par-6 family cell polarity regulator beta |
| chr21_-_18985158 | 0.04 |
ENST00000339775.6
|
BTG3
|
BTG family, member 3 |
| chr6_+_134274322 | 0.04 |
ENST00000367871.1
ENST00000237264.4 |
TBPL1
|
TBP-like 1 |
| chr5_+_96271141 | 0.04 |
ENST00000231368.5
|
LNPEP
|
leucyl/cystinyl aminopeptidase |
| chr18_-_29522989 | 0.04 |
ENST00000582539.1
ENST00000283351.4 ENST00000582513.1 |
TRAPPC8
|
trafficking protein particle complex 8 |
| chr13_-_77601282 | 0.04 |
ENST00000355619.5
|
FBXL3
|
F-box and leucine-rich repeat protein 3 |
| chr8_+_8860314 | 0.04 |
ENST00000250263.7
ENST00000519292.1 |
ERI1
|
exoribonuclease 1 |
| chr9_+_91003271 | 0.03 |
ENST00000375859.3
ENST00000541629.1 |
SPIN1
|
spindlin 1 |
| chr2_-_9143786 | 0.03 |
ENST00000462696.1
ENST00000305997.3 |
MBOAT2
|
membrane bound O-acyltransferase domain containing 2 |
| chr5_-_171711061 | 0.03 |
ENST00000393792.2
|
UBTD2
|
ubiquitin domain containing 2 |
| chr12_+_12764773 | 0.03 |
ENST00000228865.2
|
CREBL2
|
cAMP responsive element binding protein-like 2 |
| chr9_-_74383799 | 0.03 |
ENST00000377044.4
|
TMEM2
|
transmembrane protein 2 |
| chr20_+_56884752 | 0.03 |
ENST00000244040.3
|
RAB22A
|
RAB22A, member RAS oncogene family |
| chr18_-_45456930 | 0.03 |
ENST00000262160.6
ENST00000587269.1 |
SMAD2
|
SMAD family member 2 |
| chr12_+_69864129 | 0.03 |
ENST00000547219.1
ENST00000299293.2 ENST00000549921.1 ENST00000550316.1 ENST00000548154.1 ENST00000547414.1 ENST00000550389.1 ENST00000550937.1 ENST00000549092.1 ENST00000550169.1 |
FRS2
|
fibroblast growth factor receptor substrate 2 |
| chr5_-_78809950 | 0.03 |
ENST00000334082.6
|
HOMER1
|
homer homolog 1 (Drosophila) |
| chr12_-_44200052 | 0.03 |
ENST00000548315.1
ENST00000552521.1 ENST00000546662.1 ENST00000548403.1 ENST00000546506.1 |
TWF1
|
twinfilin actin-binding protein 1 |
| chr21_-_40685477 | 0.03 |
ENST00000342449.3
|
BRWD1
|
bromodomain and WD repeat domain containing 1 |
| chr3_-_169899504 | 0.03 |
ENST00000474275.1
ENST00000484931.1 ENST00000494943.1 ENST00000497658.1 ENST00000465896.1 ENST00000475729.1 ENST00000495893.2 ENST00000481639.1 ENST00000467570.1 ENST00000466189.1 |
PHC3
|
polyhomeotic homolog 3 (Drosophila) |
| chr18_+_20513782 | 0.03 |
ENST00000399722.2
ENST00000399725.2 ENST00000399721.2 ENST00000583594.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr9_+_15553055 | 0.03 |
ENST00000380701.3
|
CCDC171
|
coiled-coil domain containing 171 |
| chrX_+_108780062 | 0.02 |
ENST00000372106.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr1_+_22379120 | 0.02 |
ENST00000400259.1
ENST00000344548.3 |
CDC42
|
cell division cycle 42 |
| chrX_-_24045303 | 0.02 |
ENST00000328046.8
|
KLHL15
|
kelch-like family member 15 |
| chr4_+_6784401 | 0.02 |
ENST00000425103.1
ENST00000307659.5 |
KIAA0232
|
KIAA0232 |
| chr10_-_50323543 | 0.02 |
ENST00000332853.4
ENST00000298454.3 |
VSTM4
|
V-set and transmembrane domain containing 4 |
| chr1_-_55680762 | 0.02 |
ENST00000407756.1
ENST00000294383.6 |
USP24
|
ubiquitin specific peptidase 24 |
| chr1_+_173684047 | 0.02 |
ENST00000546011.1
ENST00000209884.4 |
KLHL20
|
kelch-like family member 20 |
| chr6_-_111136513 | 0.02 |
ENST00000368911.3
|
CDK19
|
cyclin-dependent kinase 19 |
| chr5_+_108083517 | 0.02 |
ENST00000281092.4
ENST00000536402.1 |
FER
|
fer (fps/fes related) tyrosine kinase |
| chrX_+_13707235 | 0.02 |
ENST00000464506.1
|
RAB9A
|
RAB9A, member RAS oncogene family |
| chr4_-_157892498 | 0.02 |
ENST00000502773.1
|
PDGFC
|
platelet derived growth factor C |
| chr10_-_71930222 | 0.02 |
ENST00000458634.2
ENST00000373239.2 ENST00000373242.2 ENST00000373241.4 |
SAR1A
|
SAR1 homolog A (S. cerevisiae) |
| chr14_+_103058948 | 0.02 |
ENST00000262241.6
|
RCOR1
|
REST corepressor 1 |
| chr6_-_86352642 | 0.02 |
ENST00000355238.6
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chrX_-_80065146 | 0.02 |
ENST00000373275.4
|
BRWD3
|
bromodomain and WD repeat domain containing 3 |
| chr6_+_16238786 | 0.02 |
ENST00000259727.4
|
GMPR
|
guanosine monophosphate reductase |
| chr3_+_141205852 | 0.02 |
ENST00000286364.3
ENST00000452898.1 |
RASA2
|
RAS p21 protein activator 2 |
| chr14_+_52456327 | 0.02 |
ENST00000556760.1
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr2_+_173940442 | 0.02 |
ENST00000409176.2
ENST00000338983.3 ENST00000431503.2 |
MLTK
|
Mitogen-activated protein kinase kinase kinase MLT |
| chr7_-_44924939 | 0.02 |
ENST00000395699.2
|
PURB
|
purine-rich element binding protein B |
| chr17_+_27717415 | 0.01 |
ENST00000583121.1
ENST00000261716.3 |
TAOK1
|
TAO kinase 1 |
| chr2_-_201828356 | 0.01 |
ENST00000234296.2
|
ORC2
|
origin recognition complex, subunit 2 |
| chr1_+_100435315 | 0.01 |
ENST00000370155.3
ENST00000465289.1 |
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
| chr16_-_57219966 | 0.01 |
ENST00000565760.1
ENST00000309137.8 ENST00000570184.1 ENST00000562324.1 |
FAM192A
|
family with sequence similarity 192, member A |
| chr4_+_55524085 | 0.01 |
ENST00000412167.2
ENST00000288135.5 |
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
| chr10_-_98273668 | 0.01 |
ENST00000357947.3
|
TLL2
|
tolloid-like 2 |
| chr1_+_78245303 | 0.01 |
ENST00000370791.3
ENST00000443751.2 |
FAM73A
|
family with sequence similarity 73, member A |
| chr5_-_142783175 | 0.01 |
ENST00000231509.3
ENST00000394464.2 |
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr1_+_167190066 | 0.01 |
ENST00000367866.2
ENST00000429375.2 ENST00000452019.1 ENST00000420254.3 ENST00000541643.3 |
POU2F1
|
POU class 2 homeobox 1 |
| chr15_-_34628951 | 0.01 |
ENST00000397707.2
ENST00000560611.1 |
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr14_-_81687197 | 0.01 |
ENST00000553612.1
|
GTF2A1
|
general transcription factor IIA, 1, 19/37kDa |
| chr4_+_154125565 | 0.01 |
ENST00000338700.5
|
TRIM2
|
tripartite motif containing 2 |
| chr12_+_109535373 | 0.01 |
ENST00000242576.2
|
UNG
|
uracil-DNA glycosylase |
| chr1_+_62208091 | 0.01 |
ENST00000316485.6
ENST00000371158.2 |
INADL
|
InaD-like (Drosophila) |
| chr8_+_28747884 | 0.01 |
ENST00000287701.10
ENST00000444075.1 ENST00000403668.2 ENST00000519662.1 ENST00000558662.1 ENST00000523613.1 ENST00000560599.1 ENST00000397358.3 |
HMBOX1
|
homeobox containing 1 |
| chr8_-_18871159 | 0.01 |
ENST00000327040.8
ENST00000440756.2 |
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr6_+_112408768 | 0.01 |
ENST00000368656.2
ENST00000604268.1 |
FAM229B
|
family with sequence similarity 229, member B |
| chr3_+_105085734 | 0.01 |
ENST00000306107.5
|
ALCAM
|
activated leukocyte cell adhesion molecule |
| chr12_+_56360550 | 0.01 |
ENST00000266970.4
|
CDK2
|
cyclin-dependent kinase 2 |
| chr12_-_117319236 | 0.01 |
ENST00000257572.5
|
HRK
|
harakiri, BCL2 interacting protein (contains only BH3 domain) |
| chr7_-_111846435 | 0.01 |
ENST00000437633.1
ENST00000428084.1 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr17_+_68165657 | 0.01 |
ENST00000243457.3
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr1_+_42846443 | 0.01 |
ENST00000410070.2
ENST00000431473.3 |
RIMKLA
|
ribosomal modification protein rimK-like family member A |
| chr3_+_43328004 | 0.01 |
ENST00000454177.1
ENST00000429705.2 ENST00000296088.7 ENST00000437827.1 |
SNRK
|
SNF related kinase |
| chr4_+_166794383 | 0.01 |
ENST00000061240.2
ENST00000507499.1 |
TLL1
|
tolloid-like 1 |
| chr9_-_127952032 | 0.01 |
ENST00000456642.1
ENST00000373546.3 ENST00000373547.4 |
PPP6C
|
protein phosphatase 6, catalytic subunit |
| chr1_-_20812690 | 0.01 |
ENST00000375078.3
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
| chr12_+_54943134 | 0.01 |
ENST00000243052.3
|
PDE1B
|
phosphodiesterase 1B, calmodulin-dependent |
| chr2_+_24714729 | 0.01 |
ENST00000406961.1
ENST00000405141.1 |
NCOA1
|
nuclear receptor coactivator 1 |
| chr2_-_26101374 | 0.01 |
ENST00000435504.4
|
ASXL2
|
additional sex combs like 2 (Drosophila) |
| chr20_+_43104508 | 0.01 |
ENST00000262605.4
ENST00000372904.3 |
TTPAL
|
tocopherol (alpha) transfer protein-like |
| chr3_-_33481835 | 0.01 |
ENST00000283629.3
|
UBP1
|
upstream binding protein 1 (LBP-1a) |
| chr11_-_79151695 | 0.01 |
ENST00000278550.7
|
TENM4
|
teneurin transmembrane protein 4 |
| chr4_+_79697495 | 0.01 |
ENST00000502871.1
ENST00000335016.5 |
BMP2K
|
BMP2 inducible kinase |
| chr8_+_65492756 | 0.01 |
ENST00000321870.1
|
BHLHE22
|
basic helix-loop-helix family, member e22 |
| chr4_+_25314388 | 0.01 |
ENST00000302874.4
|
ZCCHC4
|
zinc finger, CCHC domain containing 4 |
| chr11_+_94822968 | 0.01 |
ENST00000278505.4
|
ENDOD1
|
endonuclease domain containing 1 |
| chr15_+_76629064 | 0.00 |
ENST00000290759.4
|
ISL2
|
ISL LIM homeobox 2 |
| chr3_+_37903432 | 0.00 |
ENST00000443503.2
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr12_+_94542459 | 0.00 |
ENST00000258526.4
|
PLXNC1
|
plexin C1 |
| chr5_+_125935960 | 0.00 |
ENST00000297540.4
|
PHAX
|
phosphorylated adaptor for RNA export |
| chr6_+_39760783 | 0.00 |
ENST00000398904.2
ENST00000538976.1 |
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr5_+_31193847 | 0.00 |
ENST00000514738.1
ENST00000265071.2 |
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney) |
| chr2_+_12857015 | 0.00 |
ENST00000155926.4
|
TRIB2
|
tribbles pseudokinase 2 |
| chr14_+_55518349 | 0.00 |
ENST00000395468.4
|
MAPK1IP1L
|
mitogen-activated protein kinase 1 interacting protein 1-like |
| chr1_-_110052302 | 0.00 |
ENST00000369864.4
ENST00000369862.1 |
AMIGO1
|
adhesion molecule with Ig-like domain 1 |
| chr5_-_169407744 | 0.00 |
ENST00000377365.3
|
FAM196B
|
family with sequence similarity 196, member B |
| chr1_+_201617450 | 0.00 |
ENST00000295624.6
ENST00000367297.4 ENST00000367300.3 |
NAV1
|
neuron navigator 1 |
| chr6_-_53409890 | 0.00 |
ENST00000229416.6
|
GCLC
|
glutamate-cysteine ligase, catalytic subunit |
| chr19_-_46000251 | 0.00 |
ENST00000590526.1
ENST00000344680.4 ENST00000245923.4 |
RTN2
|
reticulon 2 |
| chr1_-_109506036 | 0.00 |
ENST00000369976.1
ENST00000356970.2 ENST00000369971.2 ENST00000415331.1 ENST00000357393.4 |
CLCC1
AKNAD1
|
chloride channel CLIC-like 1 AKNA domain containing 1 |
| chr7_+_129074266 | 0.00 |
ENST00000249344.2
ENST00000435494.2 |
STRIP2
|
striatin interacting protein 2 |
| chr4_-_42659102 | 0.00 |
ENST00000264449.10
ENST00000510289.1 ENST00000381668.5 |
ATP8A1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of AAGGUGC
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1904882 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.0 | GO:0036215 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.0 | 0.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |