Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
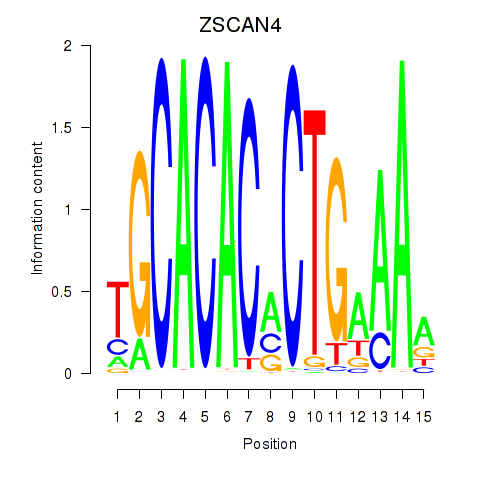
Results for ZSCAN4
Z-value: 0.82
Transcription factors associated with ZSCAN4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZSCAN4
|
ENSG00000180532.6 | zinc finger and SCAN domain containing 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZSCAN4 | hg19_v2_chr19_+_58180303_58180303 | 0.75 | 2.5e-01 | Click! |
Activity profile of ZSCAN4 motif
Sorted Z-values of ZSCAN4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_75062730 | 1.37 |
ENST00000420843.2
ENST00000360025.3 |
ARRB1
|
arrestin, beta 1 |
| chr17_-_7080227 | 1.00 |
ENST00000574330.1
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr19_+_17516494 | 0.80 |
ENST00000534306.1
|
CTD-2521M24.9
|
CTD-2521M24.9 |
| chr19_+_17516624 | 0.75 |
ENST00000596322.1
ENST00000600008.1 ENST00000601885.1 |
CTD-2521M24.9
|
CTD-2521M24.9 |
| chr15_+_78632666 | 0.72 |
ENST00000299529.6
|
CRABP1
|
cellular retinoic acid binding protein 1 |
| chr5_+_31193847 | 0.67 |
ENST00000514738.1
ENST00000265071.2 |
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney) |
| chr7_-_50633078 | 0.60 |
ENST00000444124.2
|
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase) |
| chr16_+_29690358 | 0.57 |
ENST00000395384.4
ENST00000562473.1 |
QPRT
|
quinolinate phosphoribosyltransferase |
| chr1_-_21978312 | 0.57 |
ENST00000359708.4
ENST00000290101.4 |
RAP1GAP
|
RAP1 GTPase activating protein |
| chr2_-_36779411 | 0.57 |
ENST00000406220.1
|
AC007401.2
|
Uncharacterized protein |
| chr10_+_43932553 | 0.51 |
ENST00000456416.1
ENST00000437590.2 ENST00000451167.1 |
ZNF487
|
zinc finger protein 487 |
| chr19_+_17516531 | 0.49 |
ENST00000528911.1
ENST00000528604.1 ENST00000595892.1 ENST00000500836.2 ENST00000598546.1 ENST00000600369.1 ENST00000598356.1 ENST00000594426.1 |
MVB12A
CTD-2521M24.9
|
multivesicular body subunit 12A CTD-2521M24.9 |
| chr1_-_224216371 | 0.37 |
ENST00000600307.1
|
AC138393.1
|
Uncharacterized protein |
| chr18_+_56532100 | 0.36 |
ENST00000588456.1
ENST00000589481.1 ENST00000591049.1 |
ZNF532
|
zinc finger protein 532 |
| chr14_+_53173890 | 0.34 |
ENST00000445930.2
|
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr2_-_239198743 | 0.34 |
ENST00000440245.1
ENST00000431832.1 |
PER2
|
period circadian clock 2 |
| chr6_-_153304697 | 0.33 |
ENST00000367241.3
|
FBXO5
|
F-box protein 5 |
| chr1_+_94883991 | 0.33 |
ENST00000370214.4
|
ABCD3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr14_-_55658252 | 0.32 |
ENST00000395425.2
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr11_-_33774944 | 0.32 |
ENST00000532057.1
ENST00000531080.1 |
FBXO3
|
F-box protein 3 |
| chr19_+_7660716 | 0.31 |
ENST00000160298.4
ENST00000446248.2 |
CAMSAP3
|
calmodulin regulated spectrin-associated protein family, member 3 |
| chr17_+_63133587 | 0.31 |
ENST00000449996.3
ENST00000262406.9 |
RGS9
|
regulator of G-protein signaling 9 |
| chr15_+_32885657 | 0.31 |
ENST00000448387.2
ENST00000569659.1 |
GOLGA8N
|
golgin A8 family, member N |
| chr8_+_38831683 | 0.31 |
ENST00000302495.4
|
HTRA4
|
HtrA serine peptidase 4 |
| chr21_-_44035168 | 0.30 |
ENST00000419628.1
|
AP001626.1
|
AP001626.1 |
| chr2_+_102759199 | 0.30 |
ENST00000409288.1
ENST00000410023.1 |
IL1R1
|
interleukin 1 receptor, type I |
| chr18_+_56531584 | 0.30 |
ENST00000590287.1
|
ZNF532
|
zinc finger protein 532 |
| chr11_-_1619524 | 0.29 |
ENST00000412090.1
|
KRTAP5-2
|
keratin associated protein 5-2 |
| chr2_+_237994519 | 0.28 |
ENST00000392008.2
ENST00000409334.1 ENST00000409629.1 |
COPS8
|
COP9 signalosome subunit 8 |
| chr19_-_35626104 | 0.28 |
ENST00000310123.3
ENST00000392225.3 |
LGI4
|
leucine-rich repeat LGI family, member 4 |
| chr1_-_153588765 | 0.28 |
ENST00000368701.1
ENST00000344616.2 |
S100A14
|
S100 calcium binding protein A14 |
| chr2_+_128293323 | 0.28 |
ENST00000389524.4
ENST00000428314.1 |
MYO7B
|
myosin VIIB |
| chr3_+_52009110 | 0.28 |
ENST00000491470.1
|
ABHD14A
|
abhydrolase domain containing 14A |
| chr8_+_72755796 | 0.27 |
ENST00000524152.1
|
RP11-383H13.1
|
Protein LOC100132891; cDNA FLJ53548 |
| chr12_+_133613878 | 0.27 |
ENST00000392319.2
ENST00000543758.1 |
ZNF84
|
zinc finger protein 84 |
| chr1_+_94883931 | 0.27 |
ENST00000394233.2
ENST00000454898.2 ENST00000536817.1 |
ABCD3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr17_-_33759509 | 0.27 |
ENST00000304905.5
|
SLFN12
|
schlafen family member 12 |
| chr2_+_220436917 | 0.26 |
ENST00000243786.2
|
INHA
|
inhibin, alpha |
| chr1_-_193075180 | 0.25 |
ENST00000367440.3
|
GLRX2
|
glutaredoxin 2 |
| chr9_+_128510454 | 0.24 |
ENST00000491787.3
ENST00000447726.2 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr16_+_16481306 | 0.24 |
ENST00000422673.2
|
NPIPA7
|
nuclear pore complex interacting protein family, member A7 |
| chr12_+_41086215 | 0.23 |
ENST00000547702.1
ENST00000551424.1 |
CNTN1
|
contactin 1 |
| chr12_-_81331697 | 0.23 |
ENST00000552864.1
|
LIN7A
|
lin-7 homolog A (C. elegans) |
| chr19_+_58331087 | 0.23 |
ENST00000597134.1
ENST00000593873.1 ENST00000596498.2 ENST00000603271.1 ENST00000598031.1 ENST00000316462.4 ENST00000594328.1 |
CTD-2583A14.10
ZNF587B
|
Uncharacterized protein zinc finger protein 587B |
| chrX_+_119495934 | 0.23 |
ENST00000218008.3
ENST00000361319.3 ENST00000539306.1 |
ATP1B4
|
ATPase, Na+/K+ transporting, beta 4 polypeptide |
| chr2_-_160473114 | 0.22 |
ENST00000392783.2
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr7_-_84569561 | 0.22 |
ENST00000439105.1
|
AC074183.4
|
AC074183.4 |
| chr12_-_86230315 | 0.21 |
ENST00000361228.3
|
RASSF9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chr15_-_37392703 | 0.21 |
ENST00000382766.2
ENST00000444725.1 |
MEIS2
|
Meis homeobox 2 |
| chr11_+_66624527 | 0.21 |
ENST00000393952.3
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr1_-_169337176 | 0.20 |
ENST00000472647.1
ENST00000367811.3 |
NME7
|
NME/NM23 family member 7 |
| chr12_+_122459757 | 0.20 |
ENST00000261822.4
|
BCL7A
|
B-cell CLL/lymphoma 7A |
| chr11_+_45792967 | 0.20 |
ENST00000378779.2
|
CTD-2210P24.4
|
Uncharacterized protein |
| chr14_+_53173910 | 0.19 |
ENST00000606149.1
ENST00000555339.1 ENST00000556813.1 |
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr9_+_128509624 | 0.18 |
ENST00000342287.5
ENST00000373487.4 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr19_-_11266471 | 0.18 |
ENST00000592540.1
|
SPC24
|
SPC24, NDC80 kinetochore complex component |
| chr15_-_74374891 | 0.18 |
ENST00000290438.3
|
GOLGA6A
|
golgin A6 family, member A |
| chr1_+_233765353 | 0.17 |
ENST00000366620.1
|
KCNK1
|
potassium channel, subfamily K, member 1 |
| chr5_+_176784837 | 0.17 |
ENST00000408923.3
|
RGS14
|
regulator of G-protein signaling 14 |
| chr20_+_57875658 | 0.17 |
ENST00000371025.3
|
EDN3
|
endothelin 3 |
| chr3_+_98482175 | 0.17 |
ENST00000485391.1
ENST00000492254.1 |
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr13_-_99404875 | 0.17 |
ENST00000376503.5
|
SLC15A1
|
solute carrier family 15 (oligopeptide transporter), member 1 |
| chr12_-_81331460 | 0.16 |
ENST00000549417.1
|
LIN7A
|
lin-7 homolog A (C. elegans) |
| chr22_+_24030321 | 0.16 |
ENST00000401461.1
|
RGL4
|
ral guanine nucleotide dissociation stimulator-like 4 |
| chr11_+_130029457 | 0.15 |
ENST00000278742.5
|
ST14
|
suppression of tumorigenicity 14 (colon carcinoma) |
| chr12_+_66218212 | 0.15 |
ENST00000393578.3
ENST00000425208.2 ENST00000536545.1 ENST00000354636.3 |
HMGA2
|
high mobility group AT-hook 2 |
| chr3_-_52569023 | 0.15 |
ENST00000307076.4
|
NT5DC2
|
5'-nucleotidase domain containing 2 |
| chr17_-_19648683 | 0.15 |
ENST00000573368.1
ENST00000457500.2 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr13_+_60971427 | 0.15 |
ENST00000535286.1
ENST00000377881.2 |
TDRD3
|
tudor domain containing 3 |
| chr9_+_124048864 | 0.14 |
ENST00000545652.1
|
GSN
|
gelsolin |
| chr3_-_53290016 | 0.14 |
ENST00000423525.2
ENST00000423516.1 ENST00000296289.6 ENST00000462138.1 |
TKT
|
transketolase |
| chr13_+_28813645 | 0.14 |
ENST00000282391.5
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr11_+_34643600 | 0.14 |
ENST00000530286.1
ENST00000533754.1 |
EHF
|
ets homologous factor |
| chr3_+_111393501 | 0.13 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr14_-_23284703 | 0.13 |
ENST00000555911.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr12_+_16064106 | 0.13 |
ENST00000428559.2
|
DERA
|
deoxyribose-phosphate aldolase (putative) |
| chr10_-_101989315 | 0.13 |
ENST00000370397.7
|
CHUK
|
conserved helix-loop-helix ubiquitous kinase |
| chr2_+_240323439 | 0.13 |
ENST00000428471.1
ENST00000413029.1 |
AC062017.1
|
Uncharacterized protein |
| chr21_-_47613673 | 0.13 |
ENST00000594486.1
|
AP001468.1
|
Protein LOC101060037 |
| chr15_-_37392724 | 0.13 |
ENST00000424352.2
|
MEIS2
|
Meis homeobox 2 |
| chr15_+_76135622 | 0.12 |
ENST00000338677.4
ENST00000267938.4 ENST00000569423.1 |
UBE2Q2
|
ubiquitin-conjugating enzyme E2Q family member 2 |
| chr18_+_42260059 | 0.12 |
ENST00000426838.4
|
SETBP1
|
SET binding protein 1 |
| chr1_+_15764931 | 0.12 |
ENST00000375949.4
ENST00000375943.2 |
CTRC
|
chymotrypsin C (caldecrin) |
| chr7_-_148823387 | 0.12 |
ENST00000483014.1
ENST00000378061.2 |
ZNF425
|
zinc finger protein 425 |
| chr2_-_167232484 | 0.12 |
ENST00000375387.4
ENST00000303354.6 ENST00000409672.1 |
SCN9A
|
sodium channel, voltage-gated, type IX, alpha subunit |
| chr2_+_189156389 | 0.12 |
ENST00000409843.1
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr17_-_19648916 | 0.12 |
ENST00000444455.1
ENST00000439102.2 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr7_-_135194822 | 0.11 |
ENST00000428680.2
ENST00000315544.5 ENST00000423368.2 ENST00000451834.1 ENST00000361528.4 ENST00000356162.4 ENST00000541284.1 |
CNOT4
|
CCR4-NOT transcription complex, subunit 4 |
| chr6_-_41909466 | 0.11 |
ENST00000414200.2
|
CCND3
|
cyclin D3 |
| chr19_-_42636543 | 0.11 |
ENST00000528894.4
ENST00000560804.2 ENST00000560558.1 ENST00000560398.1 ENST00000526816.2 |
POU2F2
|
POU class 2 homeobox 2 |
| chr9_+_103189405 | 0.10 |
ENST00000395067.2
|
MSANTD3
|
Myb/SANT-like DNA-binding domain containing 3 |
| chr1_+_52195542 | 0.10 |
ENST00000462759.1
ENST00000486942.1 |
OSBPL9
|
oxysterol binding protein-like 9 |
| chr19_-_42636617 | 0.10 |
ENST00000529067.1
ENST00000529952.1 ENST00000533720.1 ENST00000389341.5 ENST00000342301.4 |
POU2F2
|
POU class 2 homeobox 2 |
| chr8_+_85095553 | 0.09 |
ENST00000521268.1
|
RALYL
|
RALY RNA binding protein-like |
| chr14_+_89290965 | 0.09 |
ENST00000345383.5
ENST00000536576.1 ENST00000346301.4 ENST00000338104.6 ENST00000354441.6 ENST00000380656.2 ENST00000556651.1 ENST00000554686.1 |
TTC8
|
tetratricopeptide repeat domain 8 |
| chr3_+_50284321 | 0.09 |
ENST00000451956.1
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 |
| chr11_-_128812744 | 0.09 |
ENST00000458238.2
ENST00000531399.1 ENST00000602346.1 |
TP53AIP1
|
tumor protein p53 regulated apoptosis inducing protein 1 |
| chr7_+_27282319 | 0.09 |
ENST00000222761.3
|
EVX1
|
even-skipped homeobox 1 |
| chr15_+_28623784 | 0.09 |
ENST00000526619.2
ENST00000337838.7 ENST00000532622.2 |
GOLGA8F
|
golgin A8 family, member F |
| chr1_-_231376867 | 0.09 |
ENST00000366649.2
ENST00000318906.2 ENST00000366651.3 |
C1orf131
|
chromosome 1 open reading frame 131 |
| chr1_+_231376941 | 0.09 |
ENST00000436239.1
ENST00000366647.4 ENST00000366646.3 ENST00000416000.1 |
GNPAT
|
glyceronephosphate O-acyltransferase |
| chr15_-_28957469 | 0.08 |
ENST00000563027.1
ENST00000340249.3 |
GOLGA8M
|
golgin A8 family, member M |
| chr17_+_30348024 | 0.08 |
ENST00000327564.7
ENST00000584368.1 ENST00000394713.3 ENST00000341671.7 |
LRRC37B
|
leucine rich repeat containing 37B |
| chr17_-_19648416 | 0.08 |
ENST00000426645.2
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr17_-_58499698 | 0.07 |
ENST00000590297.1
|
USP32
|
ubiquitin specific peptidase 32 |
| chr9_+_103790991 | 0.07 |
ENST00000374874.3
|
LPPR1
|
Lipid phosphate phosphatase-related protein type 1 |
| chr6_-_41909561 | 0.07 |
ENST00000372991.4
|
CCND3
|
cyclin D3 |
| chr4_-_152682129 | 0.07 |
ENST00000512306.1
ENST00000508611.1 ENST00000515812.1 ENST00000263985.6 |
PET112
|
PET112 homolog (yeast) |
| chr11_+_111807863 | 0.07 |
ENST00000440460.2
|
DIXDC1
|
DIX domain containing 1 |
| chr14_-_21493884 | 0.07 |
ENST00000556974.1
ENST00000554419.1 ENST00000298687.5 ENST00000397858.1 ENST00000360463.3 ENST00000350792.3 ENST00000397847.2 |
NDRG2
|
NDRG family member 2 |
| chr12_-_54694807 | 0.07 |
ENST00000435572.2
|
NFE2
|
nuclear factor, erythroid 2 |
| chr19_-_40596828 | 0.07 |
ENST00000414720.2
ENST00000455521.1 ENST00000340963.5 ENST00000595773.1 |
ZNF780A
|
zinc finger protein 780A |
| chrX_+_103294483 | 0.07 |
ENST00000355016.3
|
H2BFM
|
H2B histone family, member M |
| chr5_+_127039075 | 0.06 |
ENST00000514853.2
|
CTC-228N24.1
|
CTC-228N24.1 |
| chr6_-_76203345 | 0.06 |
ENST00000393004.2
|
FILIP1
|
filamin A interacting protein 1 |
| chr2_-_101034070 | 0.06 |
ENST00000264249.3
|
CHST10
|
carbohydrate sulfotransferase 10 |
| chr13_-_52733670 | 0.06 |
ENST00000550841.1
|
NEK3
|
NIMA-related kinase 3 |
| chr10_-_103880209 | 0.06 |
ENST00000425280.1
|
LDB1
|
LIM domain binding 1 |
| chr2_-_27603582 | 0.06 |
ENST00000323703.6
ENST00000436006.1 |
ZNF513
|
zinc finger protein 513 |
| chr2_+_95831529 | 0.06 |
ENST00000295210.6
ENST00000453539.2 |
ZNF2
|
zinc finger protein 2 |
| chr2_+_234545092 | 0.05 |
ENST00000344644.5
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr11_+_71938925 | 0.05 |
ENST00000538751.1
|
INPPL1
|
inositol polyphosphate phosphatase-like 1 |
| chrX_-_20134990 | 0.05 |
ENST00000379651.3
ENST00000443379.3 ENST00000379643.5 |
MAP7D2
|
MAP7 domain containing 2 |
| chr8_-_72987810 | 0.05 |
ENST00000262209.4
|
TRPA1
|
transient receptor potential cation channel, subfamily A, member 1 |
| chr6_-_32083106 | 0.05 |
ENST00000442721.1
|
TNXB
|
tenascin XB |
| chr4_-_103682071 | 0.05 |
ENST00000505239.1
|
MANBA
|
mannosidase, beta A, lysosomal |
| chr21_-_33651324 | 0.05 |
ENST00000290130.3
|
MIS18A
|
MIS18 kinetochore protein A |
| chrX_-_62571220 | 0.04 |
ENST00000374884.2
|
SPIN4
|
spindlin family, member 4 |
| chr6_+_90272027 | 0.04 |
ENST00000522441.1
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr8_+_107593198 | 0.04 |
ENST00000517686.1
|
OXR1
|
oxidation resistance 1 |
| chr13_-_20080080 | 0.04 |
ENST00000400103.2
|
TPTE2
|
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
| chr3_+_130569429 | 0.04 |
ENST00000505330.1
ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr8_+_107670064 | 0.04 |
ENST00000312046.6
|
OXR1
|
oxidation resistance 1 |
| chr6_-_33548006 | 0.04 |
ENST00000374467.3
|
BAK1
|
BCL2-antagonist/killer 1 |
| chr13_+_60971080 | 0.04 |
ENST00000377894.2
|
TDRD3
|
tudor domain containing 3 |
| chr15_+_63413990 | 0.04 |
ENST00000261893.4
|
LACTB
|
lactamase, beta |
| chr19_-_11266437 | 0.03 |
ENST00000586708.1
ENST00000591396.1 ENST00000592967.1 ENST00000585486.1 ENST00000585567.1 |
SPC24
|
SPC24, NDC80 kinetochore complex component |
| chr12_-_57634475 | 0.03 |
ENST00000393825.1
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 |
| chr15_+_63414017 | 0.03 |
ENST00000413507.2
|
LACTB
|
lactamase, beta |
| chr3_+_130569592 | 0.03 |
ENST00000533801.2
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr15_+_75575176 | 0.03 |
ENST00000434739.3
|
GOLGA6D
|
golgin A6 family, member D |
| chrX_-_62571187 | 0.03 |
ENST00000335144.3
|
SPIN4
|
spindlin family, member 4 |
| chr20_-_16554078 | 0.03 |
ENST00000354981.2
ENST00000355755.3 ENST00000378003.2 ENST00000408042.1 |
KIF16B
|
kinesin family member 16B |
| chr16_+_4674787 | 0.03 |
ENST00000262370.7
|
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr17_-_75878647 | 0.03 |
ENST00000374983.2
|
FLJ45079
|
FLJ45079 |
| chr21_+_25801041 | 0.02 |
ENST00000453784.2
ENST00000423581.1 |
AP000476.1
|
AP000476.1 |
| chr13_-_114898016 | 0.02 |
ENST00000542651.1
ENST00000334062.7 |
RASA3
|
RAS p21 protein activator 3 |
| chr19_-_9649253 | 0.02 |
ENST00000593003.1
|
ZNF426
|
zinc finger protein 426 |
| chr7_-_10979750 | 0.02 |
ENST00000339600.5
|
NDUFA4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4, 9kDa |
| chr16_+_23847267 | 0.02 |
ENST00000321728.7
|
PRKCB
|
protein kinase C, beta |
| chr1_+_248031277 | 0.02 |
ENST00000537741.1
|
OR2W3
|
olfactory receptor, family 2, subfamily W, member 3 |
| chr16_+_89228757 | 0.02 |
ENST00000565008.1
|
LINC00304
|
long intergenic non-protein coding RNA 304 |
| chr14_-_21493649 | 0.02 |
ENST00000553442.1
ENST00000555869.1 ENST00000556457.1 ENST00000397844.2 ENST00000554415.1 |
NDRG2
|
NDRG family member 2 |
| chr10_+_47657581 | 0.02 |
ENST00000441923.2
|
ANTXRL
|
anthrax toxin receptor-like |
| chr7_-_84816122 | 0.02 |
ENST00000444867.1
|
SEMA3D
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr10_+_133753533 | 0.01 |
ENST00000422256.2
|
PPP2R2D
|
protein phosphatase 2, regulatory subunit B, delta |
| chr4_-_103682145 | 0.01 |
ENST00000226578.4
|
MANBA
|
mannosidase, beta A, lysosomal |
| chr2_+_183580954 | 0.01 |
ENST00000264065.7
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr19_+_30863271 | 0.01 |
ENST00000355537.3
|
ZNF536
|
zinc finger protein 536 |
| chr17_+_58499844 | 0.01 |
ENST00000269127.4
|
C17orf64
|
chromosome 17 open reading frame 64 |
| chr1_+_32645645 | 0.01 |
ENST00000373609.1
|
TXLNA
|
taxilin alpha |
| chr10_+_82009466 | 0.01 |
ENST00000356374.4
|
AL359195.1
|
Uncharacterized protein; cDNA FLJ46261 fis, clone TESTI4025062 |
| chr1_+_32645269 | 0.01 |
ENST00000373610.3
|
TXLNA
|
taxilin alpha |
| chr10_-_62332357 | 0.01 |
ENST00000503366.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr9_+_2017420 | 0.01 |
ENST00000439732.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr20_-_60573188 | 0.00 |
ENST00000474089.1
|
TAF4
|
TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa |
| chr19_+_49927006 | 0.00 |
ENST00000576655.1
|
CTD-3148I10.1
|
golgi-associated, olfactory signaling regulator |
| chr2_-_99952769 | 0.00 |
ENST00000409434.1
ENST00000434323.1 ENST00000264255.3 |
TXNDC9
|
thioredoxin domain containing 9 |
| chr19_-_3025614 | 0.00 |
ENST00000447365.2
|
TLE2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) |
| chr17_+_72462525 | 0.00 |
ENST00000360141.3
|
CD300A
|
CD300a molecule |
| chr17_-_40337470 | 0.00 |
ENST00000293330.1
|
HCRT
|
hypocretin (orexin) neuropeptide precursor |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZSCAN4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.2 | 0.6 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) isoquinoline alkaloid metabolic process(GO:0033076) |
| 0.2 | 0.6 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.2 | 0.7 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.4 | GO:0007387 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.1 | 0.3 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.1 | 0.3 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 0.5 | GO:0019075 | virus maturation(GO:0019075) |
| 0.1 | 0.3 | GO:0046882 | negative regulation of B cell differentiation(GO:0045578) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.1 | 0.6 | GO:0046874 | quinolinate metabolic process(GO:0046874) pyridine-containing compound catabolic process(GO:0072526) |
| 0.1 | 0.3 | GO:0019249 | lactate biosynthetic process(GO:0019249) regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.1 | 0.2 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.3 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.0 | 0.3 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 0.4 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.2 | GO:0031052 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.0 | 0.1 | GO:0005997 | xylulose metabolic process(GO:0005997) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.2 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.3 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.3 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.1 | GO:0044856 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.5 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.2 | GO:0042262 | DNA protection(GO:0042262) |
| 0.0 | 0.2 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0072642 | interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.0 | 0.2 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.0 | GO:0002352 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 1.1 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 1.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.5 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.2 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.4 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.1 | 0.6 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.1 | 1.0 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.1 | 0.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.2 | GO:0005427 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.0 | 0.5 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.3 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.0 | 0.3 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.2 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.0 | 0.1 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.0 | 0.7 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.0 | 0.3 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.3 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.2 | GO:0030613 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.0 | 0.4 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.0 | GO:0097604 | temperature-gated cation channel activity(GO:0097604) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.0 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.9 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |