Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
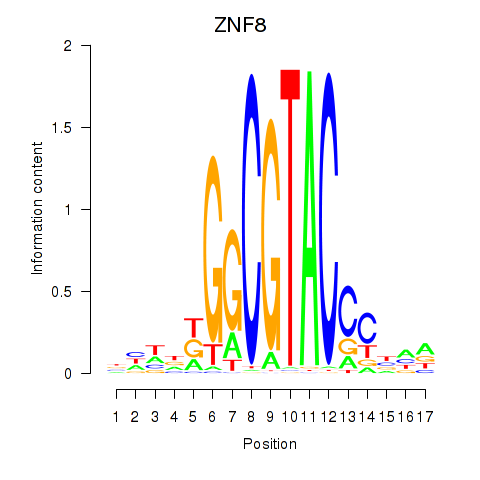
Results for ZNF8
Z-value: 1.14
Transcription factors associated with ZNF8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF8
|
ENSG00000083842.8 | zinc finger protein 8 |
|
ZNF8
|
ENSG00000273439.1 | zinc finger protein 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF8 | hg19_v2_chr19_+_58790314_58790358 | -0.66 | 3.4e-01 | Click! |
Activity profile of ZNF8 motif
Sorted Z-values of ZNF8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_56591978 | 1.03 |
ENST00000583656.1
|
MTMR4
|
myotubularin related protein 4 |
| chr6_+_29691056 | 0.96 |
ENST00000414333.1
ENST00000334668.4 ENST00000259951.7 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr17_+_48585794 | 0.94 |
ENST00000576179.1
ENST00000419930.1 |
MYCBPAP
|
MYCBP associated protein |
| chr12_-_74686314 | 0.92 |
ENST00000551210.1
ENST00000515416.2 ENST00000549905.1 |
RP11-81H3.2
|
RP11-81H3.2 |
| chr1_-_186344802 | 0.78 |
ENST00000451586.1
|
TPR
|
translocated promoter region, nuclear basket protein |
| chr12_+_104235229 | 0.67 |
ENST00000551650.1
|
RP11-650K20.3
|
Uncharacterized protein |
| chr6_+_29691198 | 0.64 |
ENST00000440587.2
ENST00000434407.2 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr2_-_240322685 | 0.55 |
ENST00000544989.1
|
HDAC4
|
histone deacetylase 4 |
| chr1_+_70671363 | 0.54 |
ENST00000370951.1
ENST00000370950.3 ENST00000405432.1 ENST00000454435.2 |
SRSF11
|
serine/arginine-rich splicing factor 11 |
| chr11_+_46402482 | 0.42 |
ENST00000441869.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr5_+_138677515 | 0.42 |
ENST00000265192.4
ENST00000511706.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr1_+_186344883 | 0.39 |
ENST00000367470.3
|
C1orf27
|
chromosome 1 open reading frame 27 |
| chr10_+_135192695 | 0.37 |
ENST00000368539.4
ENST00000278060.5 ENST00000357296.3 |
PAOX
|
polyamine oxidase (exo-N4-amino) |
| chr10_-_1034237 | 0.37 |
ENST00000381466.1
|
AL359878.1
|
Uncharacterized protein |
| chr1_+_25664408 | 0.36 |
ENST00000374358.4
|
TMEM50A
|
transmembrane protein 50A |
| chr17_-_27169745 | 0.35 |
ENST00000583307.1
ENST00000581229.1 ENST00000582266.1 ENST00000577376.1 ENST00000577682.1 ENST00000581381.1 ENST00000341217.5 ENST00000581407.1 ENST00000583522.1 |
FAM222B
|
family with sequence similarity 222, member B |
| chr3_-_46000146 | 0.35 |
ENST00000438446.1
|
FYCO1
|
FYVE and coiled-coil domain containing 1 |
| chr19_-_18392422 | 0.35 |
ENST00000252818.3
|
JUND
|
jun D proto-oncogene |
| chr11_+_69455855 | 0.35 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr17_+_48585902 | 0.34 |
ENST00000452039.1
|
MYCBPAP
|
MYCBP associated protein |
| chr8_+_61429416 | 0.33 |
ENST00000262646.7
ENST00000531289.1 |
RAB2A
|
RAB2A, member RAS oncogene family |
| chr11_+_46402583 | 0.32 |
ENST00000359803.3
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr1_+_186344945 | 0.28 |
ENST00000419367.3
ENST00000287859.6 |
C1orf27
|
chromosome 1 open reading frame 27 |
| chr3_-_9994021 | 0.26 |
ENST00000411976.2
ENST00000412055.1 |
PRRT3
|
proline-rich transmembrane protein 3 |
| chr2_-_240322643 | 0.26 |
ENST00000345617.3
|
HDAC4
|
histone deacetylase 4 |
| chr12_-_46662888 | 0.25 |
ENST00000546893.1
|
SLC38A1
|
solute carrier family 38, member 1 |
| chr5_-_43515125 | 0.25 |
ENST00000509489.1
|
C5orf34
|
chromosome 5 open reading frame 34 |
| chr9_+_96026230 | 0.24 |
ENST00000448251.1
|
WNK2
|
WNK lysine deficient protein kinase 2 |
| chr2_+_198365122 | 0.23 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough |
| chr9_-_94711007 | 0.23 |
ENST00000375715.1
|
ROR2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr9_-_32552551 | 0.22 |
ENST00000360538.2
ENST00000379858.1 |
TOPORS
|
topoisomerase I binding, arginine/serine-rich, E3 ubiquitin protein ligase |
| chr19_-_5903714 | 0.22 |
ENST00000586349.1
ENST00000585661.1 ENST00000308961.4 ENST00000592634.1 ENST00000418389.2 ENST00000252675.5 |
AC024592.12
NDUFA11
FUT5
|
Uncharacterized protein NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa fucosyltransferase 5 (alpha (1,3) fucosyltransferase) |
| chr22_+_30163340 | 0.21 |
ENST00000330029.6
ENST00000401406.3 |
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr13_+_21750780 | 0.19 |
ENST00000309594.4
|
MRP63
|
mitochondrial ribosomal protein 63 |
| chr2_+_169926047 | 0.19 |
ENST00000428522.1
ENST00000450153.1 ENST00000421653.1 |
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr17_+_75181292 | 0.19 |
ENST00000431431.2
|
SEC14L1
|
SEC14-like 1 (S. cerevisiae) |
| chr8_+_145064233 | 0.19 |
ENST00000529301.1
ENST00000395068.4 |
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr11_-_46638378 | 0.18 |
ENST00000529192.1
|
HARBI1
|
harbinger transposase derived 1 |
| chr11_+_13690200 | 0.18 |
ENST00000354817.3
|
FAR1
|
fatty acyl CoA reductase 1 |
| chr6_+_53659746 | 0.16 |
ENST00000370888.1
|
LRRC1
|
leucine rich repeat containing 1 |
| chr8_-_109260897 | 0.13 |
ENST00000521297.1
ENST00000519030.1 ENST00000521440.1 ENST00000518345.1 ENST00000519627.1 ENST00000220849.5 |
EIF3E
|
eukaryotic translation initiation factor 3, subunit E |
| chr2_+_173600514 | 0.12 |
ENST00000264111.6
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr2_+_173600565 | 0.12 |
ENST00000397081.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr13_+_53029564 | 0.11 |
ENST00000468284.1
ENST00000378034.3 ENST00000258607.5 ENST00000378037.5 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr22_+_37415676 | 0.11 |
ENST00000401419.3
|
MPST
|
mercaptopyruvate sulfurtransferase |
| chr6_+_116601265 | 0.11 |
ENST00000452085.3
|
DSE
|
dermatan sulfate epimerase |
| chr1_-_246670614 | 0.10 |
ENST00000403792.3
|
SMYD3
|
SET and MYND domain containing 3 |
| chr7_+_117824086 | 0.10 |
ENST00000249299.2
ENST00000424702.1 |
NAA38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit |
| chr5_+_10353780 | 0.10 |
ENST00000449913.2
ENST00000503788.1 ENST00000274140.5 |
MARCH6
|
membrane-associated ring finger (C3HC4) 6, E3 ubiquitin protein ligase |
| chr3_+_49236910 | 0.09 |
ENST00000452691.2
ENST00000366429.2 |
CCDC36
|
coiled-coil domain containing 36 |
| chr22_+_37415728 | 0.09 |
ENST00000404802.3
|
MPST
|
mercaptopyruvate sulfurtransferase |
| chr11_-_61560254 | 0.09 |
ENST00000543510.1
|
TMEM258
|
transmembrane protein 258 |
| chr12_+_79258444 | 0.08 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr5_+_102455968 | 0.08 |
ENST00000358359.3
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr1_-_100715372 | 0.08 |
ENST00000370131.3
ENST00000370132.4 |
DBT
|
dihydrolipoamide branched chain transacylase E2 |
| chr7_+_100303676 | 0.08 |
ENST00000303151.4
|
POP7
|
processing of precursor 7, ribonuclease P/MRP subunit (S. cerevisiae) |
| chrX_+_1387693 | 0.07 |
ENST00000381529.3
ENST00000432318.2 ENST00000361536.3 ENST00000501036.2 ENST00000381524.3 ENST00000412290.1 |
CSF2RA
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr2_+_172290707 | 0.07 |
ENST00000375255.3
ENST00000539783.1 |
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr22_+_37415776 | 0.07 |
ENST00000341116.3
ENST00000429360.2 ENST00000404393.1 |
MPST
|
mercaptopyruvate sulfurtransferase |
| chr22_-_30162924 | 0.06 |
ENST00000344318.3
ENST00000397781.3 |
ZMAT5
|
zinc finger, matrin-type 5 |
| chr10_-_13043697 | 0.06 |
ENST00000378825.3
|
CCDC3
|
coiled-coil domain containing 3 |
| chr15_+_85523671 | 0.06 |
ENST00000310298.4
ENST00000557957.1 |
PDE8A
|
phosphodiesterase 8A |
| chr14_+_51706886 | 0.05 |
ENST00000457354.2
|
TMX1
|
thioredoxin-related transmembrane protein 1 |
| chr20_+_37377085 | 0.05 |
ENST00000243903.4
|
ACTR5
|
ARP5 actin-related protein 5 homolog (yeast) |
| chr13_-_25861530 | 0.05 |
ENST00000540661.1
|
MTMR6
|
myotubularin related protein 6 |
| chr9_+_1050331 | 0.05 |
ENST00000382255.3
ENST00000382251.3 ENST00000412350.2 |
DMRT2
|
doublesex and mab-3 related transcription factor 2 |
| chr1_-_55266926 | 0.05 |
ENST00000371276.4
|
TTC22
|
tetratricopeptide repeat domain 22 |
| chr5_+_89854595 | 0.05 |
ENST00000405460.2
|
GPR98
|
G protein-coupled receptor 98 |
| chr6_+_106959718 | 0.04 |
ENST00000369066.3
|
AIM1
|
absent in melanoma 1 |
| chr7_+_1748798 | 0.04 |
ENST00000561626.1
|
ELFN1
|
extracellular leucine-rich repeat and fibronectin type III domain containing 1 |
| chr8_+_38244638 | 0.04 |
ENST00000526356.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr22_+_37415700 | 0.04 |
ENST00000397129.1
|
MPST
|
mercaptopyruvate sulfurtransferase |
| chr11_+_46638805 | 0.03 |
ENST00000434074.1
ENST00000312040.4 ENST00000451945.1 |
ATG13
|
autophagy related 13 |
| chr2_+_87144738 | 0.03 |
ENST00000559485.1
|
RGPD1
|
RANBP2-like and GRIP domain containing 1 |
| chr17_+_34136459 | 0.02 |
ENST00000588240.1
ENST00000590273.1 ENST00000588441.1 ENST00000587272.1 ENST00000592237.1 ENST00000311979.3 |
TAF15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa |
| chrX_+_128913906 | 0.01 |
ENST00000356892.3
|
SASH3
|
SAM and SH3 domain containing 3 |
| chr19_+_35739782 | 0.01 |
ENST00000347609.4
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr12_+_79258547 | 0.01 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr4_-_52904425 | 0.00 |
ENST00000535450.1
|
SGCB
|
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
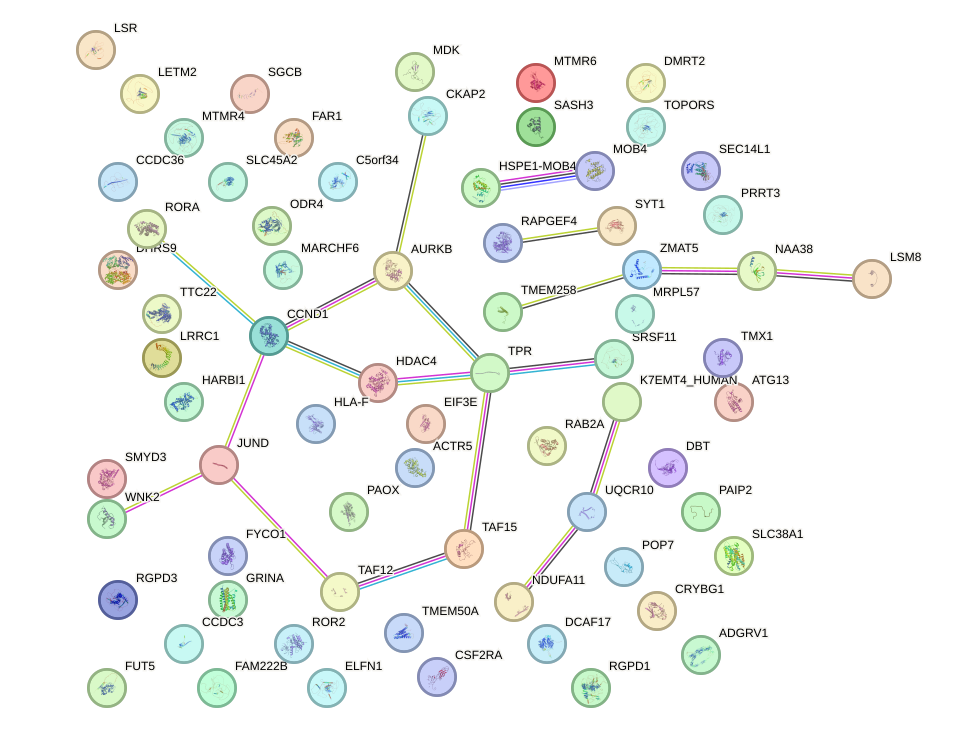
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF8
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0045799 | negative regulation of nucleobase-containing compound transport(GO:0032240) positive regulation of chromatin assembly or disassembly(GO:0045799) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.2 | 1.6 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 0.7 | GO:0030421 | defecation(GO:0030421) |
| 0.1 | 1.8 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.4 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 0.0 | 0.2 | GO:0009439 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.0 | 0.2 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.4 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.4 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.1 | GO:0060629 | meiotic DNA double-strand break formation(GO:0042138) regulation of homologous chromosome segregation(GO:0060629) |
| 0.0 | 0.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.2 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.2 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.5 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.2 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.2 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.8 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.4 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.1 | 0.8 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.1 | 0.3 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.1 | 0.4 | GO:0052901 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.1 | 1.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.2 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.2 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.2 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.8 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.2 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.2 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |