Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
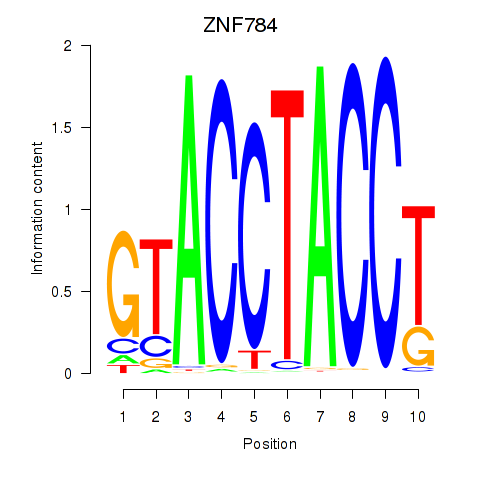
Results for ZNF784
Z-value: 0.57
Transcription factors associated with ZNF784
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF784
|
ENSG00000179922.5 | zinc finger protein 784 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF784 | hg19_v2_chr19_-_56135928_56135967 | -0.80 | 2.0e-01 | Click! |
Activity profile of ZNF784 motif
Sorted Z-values of ZNF784 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_37723336 | 0.51 |
ENST00000450180.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr2_-_191885686 | 0.39 |
ENST00000432058.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr7_+_114562616 | 0.35 |
ENST00000448022.1
|
MDFIC
|
MyoD family inhibitor domain containing |
| chr3_+_188817891 | 0.33 |
ENST00000412373.1
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr17_+_73717516 | 0.32 |
ENST00000200181.3
ENST00000339591.3 |
ITGB4
|
integrin, beta 4 |
| chr3_-_42306248 | 0.31 |
ENST00000334681.5
|
CCK
|
cholecystokinin |
| chr17_-_19281203 | 0.31 |
ENST00000487415.2
|
B9D1
|
B9 protein domain 1 |
| chr17_+_73717551 | 0.30 |
ENST00000450894.3
|
ITGB4
|
integrin, beta 4 |
| chr22_-_37640277 | 0.30 |
ENST00000401529.3
ENST00000249071.6 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr17_+_73717407 | 0.30 |
ENST00000579662.1
|
ITGB4
|
integrin, beta 4 |
| chr22_-_37640456 | 0.28 |
ENST00000405484.1
ENST00000441619.1 ENST00000406508.1 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr11_+_65154070 | 0.25 |
ENST00000317568.5
ENST00000531296.1 ENST00000533782.1 ENST00000355991.5 ENST00000416776.2 ENST00000526201.1 |
FRMD8
|
FERM domain containing 8 |
| chr1_+_55446465 | 0.25 |
ENST00000371268.3
|
TMEM61
|
transmembrane protein 61 |
| chr17_+_77020325 | 0.25 |
ENST00000311661.4
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr17_+_77020224 | 0.24 |
ENST00000339142.2
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr19_+_7562431 | 0.24 |
ENST00000361664.2
|
C19orf45
|
chromosome 19 open reading frame 45 |
| chr2_+_45878790 | 0.23 |
ENST00000306156.3
|
PRKCE
|
protein kinase C, epsilon |
| chr5_-_158526756 | 0.23 |
ENST00000313708.6
ENST00000517373.1 |
EBF1
|
early B-cell factor 1 |
| chr4_-_130692631 | 0.22 |
ENST00000500092.2
ENST00000509105.1 |
RP11-519M16.1
|
RP11-519M16.1 |
| chr14_-_25479811 | 0.22 |
ENST00000550887.1
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr6_-_150039170 | 0.22 |
ENST00000458696.2
ENST00000392273.3 |
LATS1
|
large tumor suppressor kinase 1 |
| chr11_-_115375107 | 0.22 |
ENST00000545380.1
ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1
|
cell adhesion molecule 1 |
| chr1_-_1149506 | 0.21 |
ENST00000379236.3
|
TNFRSF4
|
tumor necrosis factor receptor superfamily, member 4 |
| chr21_+_41029235 | 0.20 |
ENST00000380618.1
|
B3GALT5
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 5 |
| chr2_+_231191875 | 0.20 |
ENST00000444636.1
ENST00000415673.2 ENST00000243810.6 ENST00000396563.4 |
SP140L
|
SP140 nuclear body protein-like |
| chr19_-_10213335 | 0.20 |
ENST00000592641.1
ENST00000253109.4 |
ANGPTL6
|
angiopoietin-like 6 |
| chr3_+_9834179 | 0.20 |
ENST00000498623.2
|
ARPC4
|
actin related protein 2/3 complex, subunit 4, 20kDa |
| chr8_-_23563922 | 0.20 |
ENST00000418222.1
ENST00000325017.3 |
NKX2-6
|
NK2 homeobox 6 |
| chr5_-_137674000 | 0.20 |
ENST00000510119.1
ENST00000513970.1 |
CDC25C
|
cell division cycle 25C |
| chr15_-_74501360 | 0.19 |
ENST00000323940.5
|
STRA6
|
stimulated by retinoic acid 6 |
| chr3_-_197476560 | 0.19 |
ENST00000273582.5
|
KIAA0226
|
KIAA0226 |
| chr7_-_130418888 | 0.18 |
ENST00000310992.4
|
KLF14
|
Kruppel-like factor 14 |
| chr7_+_22766766 | 0.18 |
ENST00000426291.1
ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6
|
interleukin 6 (interferon, beta 2) |
| chr12_+_52626898 | 0.17 |
ENST00000331817.5
|
KRT7
|
keratin 7 |
| chr4_+_1340986 | 0.17 |
ENST00000511216.1
ENST00000389851.4 |
UVSSA
|
UV-stimulated scaffold protein A |
| chr15_-_74501310 | 0.17 |
ENST00000423167.2
ENST00000432245.2 |
STRA6
|
stimulated by retinoic acid 6 |
| chr18_+_21529811 | 0.17 |
ENST00000588004.1
|
LAMA3
|
laminin, alpha 3 |
| chr2_-_73869508 | 0.17 |
ENST00000272425.3
|
NAT8
|
N-acetyltransferase 8 (GCN5-related, putative) |
| chr15_-_59665062 | 0.16 |
ENST00000288235.4
|
MYO1E
|
myosin IE |
| chr11_-_8290263 | 0.16 |
ENST00000428101.2
|
LMO1
|
LIM domain only 1 (rhombotin 1) |
| chr17_+_77020146 | 0.16 |
ENST00000579760.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr5_+_75379224 | 0.16 |
ENST00000322285.7
|
SV2C
|
synaptic vesicle glycoprotein 2C |
| chr11_+_65122216 | 0.16 |
ENST00000309880.5
|
TIGD3
|
tigger transposable element derived 3 |
| chr16_-_57832004 | 0.15 |
ENST00000562503.1
|
KIFC3
|
kinesin family member C3 |
| chr1_+_32042131 | 0.15 |
ENST00000271064.7
ENST00000537531.1 |
TINAGL1
|
tubulointerstitial nephritis antigen-like 1 |
| chr22_+_23522552 | 0.15 |
ENST00000359540.3
ENST00000398512.5 |
BCR
|
breakpoint cluster region |
| chr2_-_70780770 | 0.15 |
ENST00000444975.1
ENST00000445399.1 ENST00000418333.2 |
TGFA
|
transforming growth factor, alpha |
| chr4_+_86396321 | 0.15 |
ENST00000503995.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chrX_-_54384425 | 0.15 |
ENST00000375169.3
ENST00000354646.2 |
WNK3
|
WNK lysine deficient protein kinase 3 |
| chr11_-_62389449 | 0.15 |
ENST00000534026.1
|
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr14_+_29241910 | 0.14 |
ENST00000399387.4
ENST00000552957.1 ENST00000548213.1 |
C14orf23
|
chromosome 14 open reading frame 23 |
| chr20_+_20348740 | 0.14 |
ENST00000310227.1
|
INSM1
|
insulinoma-associated 1 |
| chr22_+_19938419 | 0.14 |
ENST00000412786.1
|
COMT
|
catechol-O-methyltransferase |
| chr16_-_57831676 | 0.14 |
ENST00000465878.2
ENST00000539578.1 ENST00000561524.1 |
KIFC3
|
kinesin family member C3 |
| chr11_+_76494253 | 0.14 |
ENST00000333090.4
|
TSKU
|
tsukushi, small leucine rich proteoglycan |
| chr5_-_158526693 | 0.14 |
ENST00000380654.4
|
EBF1
|
early B-cell factor 1 |
| chr16_-_57831914 | 0.14 |
ENST00000421376.2
|
KIFC3
|
kinesin family member C3 |
| chr16_-_67190152 | 0.14 |
ENST00000486556.1
|
TRADD
|
TNFRSF1A-associated via death domain |
| chr9_-_110251836 | 0.14 |
ENST00000374672.4
|
KLF4
|
Kruppel-like factor 4 (gut) |
| chr2_-_47168850 | 0.14 |
ENST00000409207.1
|
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr8_-_87242589 | 0.14 |
ENST00000419776.2
ENST00000297524.3 |
SLC7A13
|
solute carrier family 7 (anionic amino acid transporter), member 13 |
| chr10_+_82168240 | 0.13 |
ENST00000372187.5
ENST00000372185.1 |
FAM213A
|
family with sequence similarity 213, member A |
| chr3_+_9834227 | 0.13 |
ENST00000287613.7
ENST00000397261.3 |
ARPC4
|
actin related protein 2/3 complex, subunit 4, 20kDa |
| chr11_-_62389577 | 0.13 |
ENST00000534715.1
|
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chrX_-_153095945 | 0.12 |
ENST00000164640.4
|
PDZD4
|
PDZ domain containing 4 |
| chr7_+_3340989 | 0.12 |
ENST00000404826.2
ENST00000389531.3 |
SDK1
|
sidekick cell adhesion molecule 1 |
| chr6_+_123100853 | 0.12 |
ENST00000356535.4
|
FABP7
|
fatty acid binding protein 7, brain |
| chr7_+_128379449 | 0.12 |
ENST00000479257.1
|
CALU
|
calumenin |
| chr11_-_5248294 | 0.12 |
ENST00000335295.4
|
HBB
|
hemoglobin, beta |
| chr15_+_25200108 | 0.12 |
ENST00000577949.1
ENST00000338094.6 ENST00000338327.4 ENST00000579070.1 ENST00000577565.1 |
SNURF
SNRPN
|
SNRPN upstream reading frame protein small nuclear ribonucleoprotein polypeptide N |
| chrX_-_106243294 | 0.12 |
ENST00000255495.7
|
MORC4
|
MORC family CW-type zinc finger 4 |
| chr12_-_91505608 | 0.12 |
ENST00000266718.4
|
LUM
|
lumican |
| chr20_-_56285595 | 0.12 |
ENST00000395816.3
ENST00000347215.4 |
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr15_-_70387120 | 0.11 |
ENST00000539550.1
|
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr10_-_127511790 | 0.11 |
ENST00000368797.4
ENST00000420761.1 |
UROS
|
uroporphyrinogen III synthase |
| chr17_+_19281034 | 0.11 |
ENST00000308406.5
ENST00000299612.7 |
MAPK7
|
mitogen-activated protein kinase 7 |
| chr4_-_90756769 | 0.11 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr6_+_123100620 | 0.11 |
ENST00000368444.3
|
FABP7
|
fatty acid binding protein 7, brain |
| chrX_-_53310791 | 0.11 |
ENST00000375365.2
|
IQSEC2
|
IQ motif and Sec7 domain 2 |
| chr22_+_19939026 | 0.11 |
ENST00000406520.3
|
COMT
|
catechol-O-methyltransferase |
| chr1_+_32042105 | 0.11 |
ENST00000457433.2
ENST00000441210.2 |
TINAGL1
|
tubulointerstitial nephritis antigen-like 1 |
| chr20_-_56286479 | 0.11 |
ENST00000265626.4
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr7_+_102191679 | 0.11 |
ENST00000507918.1
ENST00000432940.1 |
SPDYE2
|
speedy/RINGO cell cycle regulator family member E2 |
| chr4_+_130692778 | 0.11 |
ENST00000513875.1
ENST00000508724.1 |
RP11-422J15.1
|
RP11-422J15.1 |
| chr12_+_54402790 | 0.11 |
ENST00000040584.4
|
HOXC8
|
homeobox C8 |
| chr19_+_39936317 | 0.11 |
ENST00000598725.1
|
SUPT5H
|
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr7_-_29152509 | 0.11 |
ENST00000448959.1
|
CPVL
|
carboxypeptidase, vitellogenic-like |
| chr14_+_75894714 | 0.11 |
ENST00000559060.1
|
JDP2
|
Jun dimerization protein 2 |
| chr8_-_16859690 | 0.11 |
ENST00000180166.5
|
FGF20
|
fibroblast growth factor 20 |
| chr5_-_131630931 | 0.11 |
ENST00000431054.1
|
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chr17_-_42767092 | 0.11 |
ENST00000588687.1
|
CCDC43
|
coiled-coil domain containing 43 |
| chr3_+_32993065 | 0.11 |
ENST00000330953.5
|
CCR4
|
chemokine (C-C motif) receptor 4 |
| chr12_-_4754339 | 0.10 |
ENST00000228850.1
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr11_-_62389621 | 0.10 |
ENST00000531383.1
ENST00000265471.5 |
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr10_+_8096631 | 0.10 |
ENST00000379328.3
|
GATA3
|
GATA binding protein 3 |
| chr9_+_116037922 | 0.10 |
ENST00000374198.4
|
PRPF4
|
pre-mRNA processing factor 4 |
| chr19_+_6464502 | 0.10 |
ENST00000308243.7
|
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr19_+_39936267 | 0.10 |
ENST00000359191.6
|
SUPT5H
|
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr10_+_21823243 | 0.10 |
ENST00000307729.7
ENST00000377091.2 |
MLLT10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr22_+_42196666 | 0.10 |
ENST00000402061.3
ENST00000255784.5 |
CCDC134
|
coiled-coil domain containing 134 |
| chr20_-_6103666 | 0.10 |
ENST00000536936.1
|
FERMT1
|
fermitin family member 1 |
| chr5_+_175976324 | 0.10 |
ENST00000261944.5
|
CDHR2
|
cadherin-related family member 2 |
| chrX_-_153095813 | 0.10 |
ENST00000544474.1
|
PDZD4
|
PDZ domain containing 4 |
| chrX_-_49041242 | 0.10 |
ENST00000453382.1
ENST00000540849.1 ENST00000536904.1 ENST00000432913.1 |
PRICKLE3
|
prickle homolog 3 (Drosophila) |
| chr2_+_54683419 | 0.10 |
ENST00000356805.4
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr17_-_40428359 | 0.10 |
ENST00000293328.3
|
STAT5B
|
signal transducer and activator of transcription 5B |
| chr9_-_20621834 | 0.10 |
ENST00000429426.2
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr9_+_112852477 | 0.10 |
ENST00000480388.1
|
AKAP2
|
A kinase (PRKA) anchor protein 2 |
| chr22_+_25202232 | 0.09 |
ENST00000400358.4
ENST00000400359.4 |
SGSM1
|
small G protein signaling modulator 1 |
| chr14_-_64108125 | 0.09 |
ENST00000267522.3
|
WDR89
|
WD repeat domain 89 |
| chr19_+_39936186 | 0.09 |
ENST00000432763.2
ENST00000402194.2 ENST00000601515.1 |
SUPT5H
|
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr8_-_101118210 | 0.09 |
ENST00000360863.6
|
RGS22
|
regulator of G-protein signaling 22 |
| chr5_-_176943917 | 0.09 |
ENST00000330503.7
|
DDX41
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 41 |
| chr5_+_128430401 | 0.09 |
ENST00000514194.1
ENST00000173527.5 |
ISOC1
|
isochorismatase domain containing 1 |
| chr5_+_73109339 | 0.09 |
ENST00000296799.4
|
ARHGEF28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr9_+_132815985 | 0.09 |
ENST00000372410.3
|
GPR107
|
G protein-coupled receptor 107 |
| chr5_-_149535190 | 0.09 |
ENST00000517488.1
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide |
| chr17_-_73149921 | 0.09 |
ENST00000481647.1
ENST00000470924.1 |
HN1
|
hematological and neurological expressed 1 |
| chr4_-_28404486 | 0.09 |
ENST00000515720.1
|
RP11-180C1.1
|
Uncharacterized protein |
| chr2_-_145277640 | 0.09 |
ENST00000539609.3
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr8_+_356942 | 0.09 |
ENST00000276326.5
|
FBXO25
|
F-box protein 25 |
| chr11_+_64002292 | 0.09 |
ENST00000426086.2
|
VEGFB
|
vascular endothelial growth factor B |
| chrX_-_23761317 | 0.09 |
ENST00000492081.1
ENST00000379303.5 ENST00000336430.7 |
ACOT9
|
acyl-CoA thioesterase 9 |
| chr8_-_101117847 | 0.08 |
ENST00000523287.1
ENST00000519092.1 |
RGS22
|
regulator of G-protein signaling 22 |
| chr2_+_47168313 | 0.08 |
ENST00000319190.5
ENST00000394850.2 ENST00000536057.1 |
TTC7A
|
tetratricopeptide repeat domain 7A |
| chr11_+_65647280 | 0.08 |
ENST00000307886.3
ENST00000528419.1 ENST00000526034.1 |
CTSW
|
cathepsin W |
| chr19_-_18548921 | 0.08 |
ENST00000545187.1
ENST00000578352.1 |
ISYNA1
|
inositol-3-phosphate synthase 1 |
| chr17_-_74707037 | 0.08 |
ENST00000355797.3
ENST00000375036.2 ENST00000449428.2 |
MXRA7
|
matrix-remodelling associated 7 |
| chr3_-_112218378 | 0.08 |
ENST00000334529.5
|
BTLA
|
B and T lymphocyte associated |
| chr16_+_2564254 | 0.08 |
ENST00000565223.1
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c |
| chr9_-_112260531 | 0.08 |
ENST00000374541.2
ENST00000262539.3 |
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr11_+_89867681 | 0.08 |
ENST00000534061.1
|
NAALAD2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr17_+_75450075 | 0.08 |
ENST00000592951.1
|
SEPT9
|
septin 9 |
| chr7_+_114562909 | 0.08 |
ENST00000423503.1
ENST00000427207.1 |
MDFIC
|
MyoD family inhibitor domain containing |
| chr3_-_9834375 | 0.08 |
ENST00000343450.2
ENST00000301964.2 |
TADA3
|
transcriptional adaptor 3 |
| chr5_+_55147205 | 0.08 |
ENST00000396836.2
ENST00000396834.1 ENST00000447346.2 ENST00000359040.5 |
IL31RA
|
interleukin 31 receptor A |
| chr2_-_145277569 | 0.08 |
ENST00000303660.4
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr12_-_96390108 | 0.08 |
ENST00000538703.1
ENST00000261208.3 |
HAL
|
histidine ammonia-lyase |
| chr9_-_130693048 | 0.08 |
ENST00000388747.4
|
PIP5KL1
|
phosphatidylinositol-4-phosphate 5-kinase-like 1 |
| chr2_+_181988560 | 0.08 |
ENST00000424170.1
ENST00000435411.1 |
AC104820.2
|
AC104820.2 |
| chr10_-_127511658 | 0.07 |
ENST00000368774.1
ENST00000368778.3 |
UROS
|
uroporphyrinogen III synthase |
| chr17_+_55055466 | 0.07 |
ENST00000262288.3
ENST00000572710.1 ENST00000575395.1 |
SCPEP1
|
serine carboxypeptidase 1 |
| chr8_+_21914348 | 0.07 |
ENST00000520174.1
|
DMTN
|
dematin actin binding protein |
| chr12_+_47617284 | 0.07 |
ENST00000549630.1
ENST00000551777.1 |
PCED1B
|
PC-esterase domain containing 1B |
| chr1_+_228327943 | 0.07 |
ENST00000366726.1
ENST00000312726.4 ENST00000366728.2 ENST00000453943.1 ENST00000366723.1 ENST00000366722.1 ENST00000435153.1 ENST00000366721.1 |
GUK1
|
guanylate kinase 1 |
| chr19_+_58898627 | 0.07 |
ENST00000598098.1
ENST00000598495.1 ENST00000196551.3 ENST00000596046.1 |
RPS5
|
ribosomal protein S5 |
| chr2_-_74692473 | 0.07 |
ENST00000535045.1
ENST00000409065.1 ENST00000414701.1 ENST00000448666.1 ENST00000233616.4 ENST00000452063.2 |
MOGS
|
mannosyl-oligosaccharide glucosidase |
| chr6_-_43496605 | 0.07 |
ENST00000455285.2
|
XPO5
|
exportin 5 |
| chr2_+_47168630 | 0.07 |
ENST00000263737.6
|
TTC7A
|
tetratricopeptide repeat domain 7A |
| chr22_-_24315372 | 0.07 |
ENST00000428792.1
|
DDT
|
D-dopachrome tautomerase |
| chr10_+_28966271 | 0.07 |
ENST00000375533.3
|
BAMBI
|
BMP and activin membrane-bound inhibitor |
| chr17_+_75450146 | 0.07 |
ENST00000585929.1
ENST00000590917.1 |
SEPT9
|
septin 9 |
| chr19_-_375970 | 0.07 |
ENST00000346878.2
|
THEG
|
theg spermatid protein |
| chr2_-_175462456 | 0.07 |
ENST00000409891.1
ENST00000410117.1 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr2_-_96874553 | 0.07 |
ENST00000337288.5
ENST00000443962.1 |
STARD7
|
StAR-related lipid transfer (START) domain containing 7 |
| chr1_+_228327923 | 0.07 |
ENST00000391865.3
|
GUK1
|
guanylate kinase 1 |
| chr5_+_169010638 | 0.07 |
ENST00000265295.4
ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr12_+_122242597 | 0.06 |
ENST00000267197.5
|
SETD1B
|
SET domain containing 1B |
| chr9_+_130565487 | 0.06 |
ENST00000373225.3
ENST00000431857.1 |
FPGS
|
folylpolyglutamate synthase |
| chrX_-_106243451 | 0.06 |
ENST00000355610.4
ENST00000535534.1 |
MORC4
|
MORC family CW-type zinc finger 4 |
| chr13_+_24153488 | 0.06 |
ENST00000382258.4
ENST00000382263.3 |
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr8_-_101118185 | 0.06 |
ENST00000523437.1
|
RGS22
|
regulator of G-protein signaling 22 |
| chr5_-_174871136 | 0.06 |
ENST00000393752.2
|
DRD1
|
dopamine receptor D1 |
| chr9_+_136325149 | 0.06 |
ENST00000542192.1
|
CACFD1
|
calcium channel flower domain containing 1 |
| chr4_-_10686475 | 0.06 |
ENST00000226951.6
|
CLNK
|
cytokine-dependent hematopoietic cell linker |
| chr9_-_20622478 | 0.06 |
ENST00000355930.6
ENST00000380338.4 |
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr11_+_60614946 | 0.06 |
ENST00000545580.1
|
CCDC86
|
coiled-coil domain containing 86 |
| chr7_+_37723420 | 0.06 |
ENST00000476620.1
|
EPDR1
|
ependymin related 1 |
| chrX_-_57147748 | 0.06 |
ENST00000374910.3
|
SPIN2B
|
spindlin family, member 2B |
| chr7_+_90032821 | 0.06 |
ENST00000427904.1
|
CLDN12
|
claudin 12 |
| chr19_-_42498369 | 0.06 |
ENST00000302102.5
ENST00000545399.1 |
ATP1A3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr11_+_208815 | 0.06 |
ENST00000530889.1
|
RIC8A
|
RIC8 guanine nucleotide exchange factor A |
| chr19_-_376011 | 0.06 |
ENST00000342640.4
|
THEG
|
theg spermatid protein |
| chr17_-_1394940 | 0.06 |
ENST00000570984.2
ENST00000361007.2 |
MYO1C
|
myosin IC |
| chr11_+_64008525 | 0.06 |
ENST00000449942.2
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr7_+_128577972 | 0.06 |
ENST00000357234.5
ENST00000477535.1 ENST00000479582.1 ENST00000464557.1 ENST00000402030.2 |
IRF5
|
interferon regulatory factor 5 |
| chr7_-_22234381 | 0.06 |
ENST00000458533.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr12_-_12837423 | 0.06 |
ENST00000540510.1
|
GPR19
|
G protein-coupled receptor 19 |
| chr7_+_119913688 | 0.06 |
ENST00000331113.4
|
KCND2
|
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chrX_-_19504642 | 0.06 |
ENST00000469203.2
|
MAP3K15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr17_+_46132037 | 0.06 |
ENST00000582155.1
ENST00000583378.1 ENST00000536222.1 |
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr17_-_48546232 | 0.05 |
ENST00000258969.4
|
CHAD
|
chondroadherin |
| chr11_-_62314268 | 0.05 |
ENST00000257247.7
ENST00000531324.1 ENST00000378024.4 |
AHNAK
|
AHNAK nucleoprotein |
| chr4_+_41362796 | 0.05 |
ENST00000508501.1
ENST00000512946.1 ENST00000313860.7 ENST00000512632.1 ENST00000512820.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr5_-_178772424 | 0.05 |
ENST00000251582.7
ENST00000274609.5 |
ADAMTS2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
| chr3_-_39321512 | 0.05 |
ENST00000399220.2
|
CX3CR1
|
chemokine (C-X3-C motif) receptor 1 |
| chr7_-_97881429 | 0.05 |
ENST00000420697.1
ENST00000379795.3 ENST00000415086.1 ENST00000542604.1 ENST00000447648.2 |
TECPR1
|
tectonin beta-propeller repeat containing 1 |
| chr11_-_12030130 | 0.05 |
ENST00000450094.2
ENST00000534511.1 |
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr3_-_42623805 | 0.05 |
ENST00000456515.1
|
SEC22C
|
SEC22 vesicle trafficking protein homolog C (S. cerevisiae) |
| chr17_+_42264556 | 0.05 |
ENST00000319511.6
ENST00000589785.1 ENST00000592825.1 ENST00000589184.1 |
TMUB2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr4_-_53522703 | 0.05 |
ENST00000508499.1
|
USP46
|
ubiquitin specific peptidase 46 |
| chr22_+_31003133 | 0.05 |
ENST00000405742.3
|
TCN2
|
transcobalamin II |
| chr7_-_107643674 | 0.05 |
ENST00000222399.6
|
LAMB1
|
laminin, beta 1 |
| chr21_+_27543175 | 0.05 |
ENST00000608591.1
ENST00000609365.1 |
AP000230.1
|
AP000230.1 |
| chr1_-_161208013 | 0.05 |
ENST00000515452.1
ENST00000367983.4 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr13_-_33760216 | 0.05 |
ENST00000255486.4
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr5_+_137673945 | 0.05 |
ENST00000513056.1
ENST00000511276.1 |
FAM53C
|
family with sequence similarity 53, member C |
| chr1_-_92951607 | 0.05 |
ENST00000427103.1
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr20_-_33539655 | 0.05 |
ENST00000451957.2
|
GSS
|
glutathione synthetase |
| chr2_+_85839218 | 0.05 |
ENST00000448971.1
ENST00000442708.1 ENST00000450066.2 |
USP39
|
ubiquitin specific peptidase 39 |
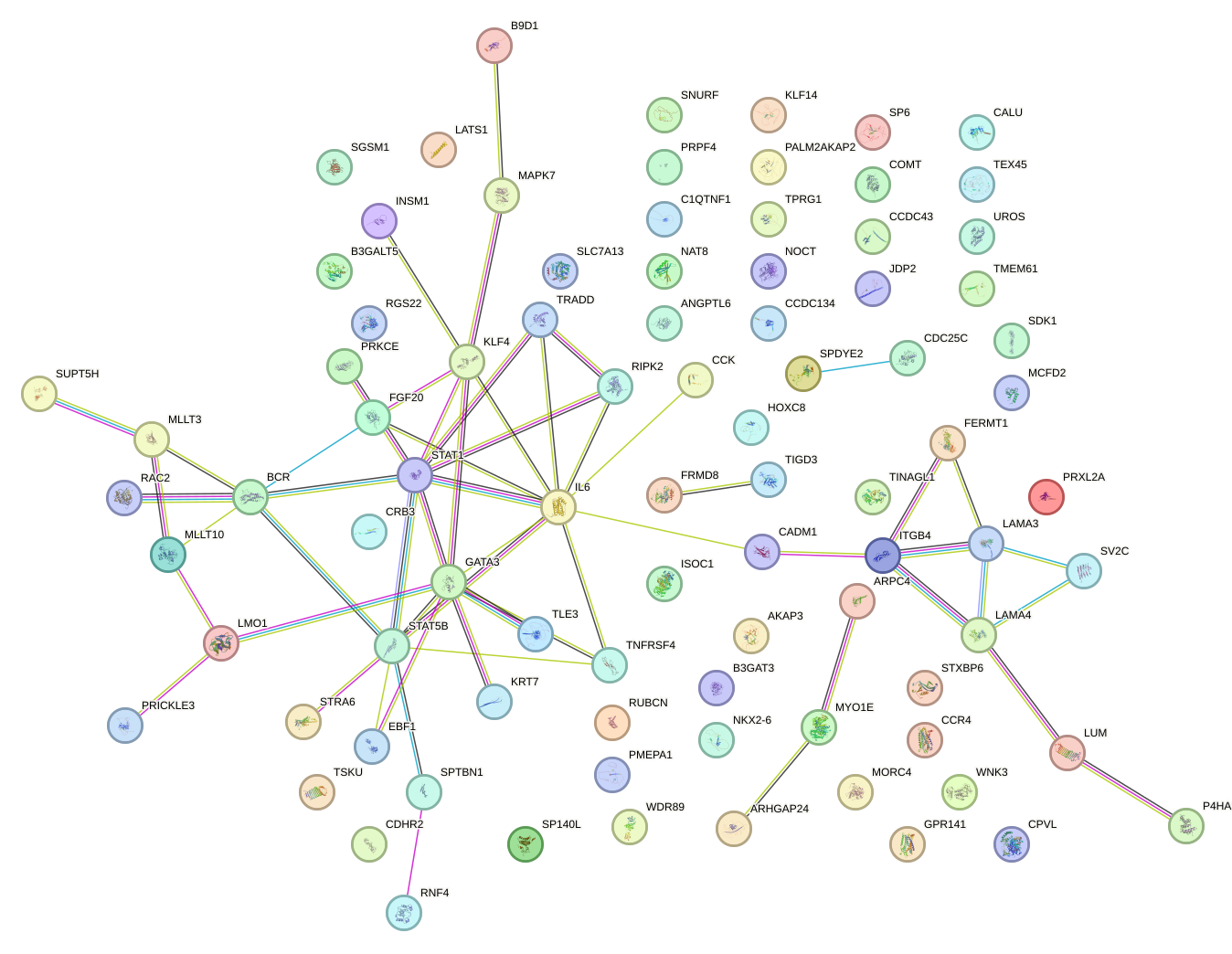
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF784
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.1 | 0.7 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.2 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.1 | 0.7 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.1 | 0.2 | GO:2000364 | regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.1 | 0.4 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.1 | 0.2 | GO:0035668 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.1 | 0.9 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 0.2 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.3 | GO:0031337 | cellular response to phosphate starvation(GO:0016036) regulation of sulfur amino acid metabolic process(GO:0031335) positive regulation of sulfur amino acid metabolic process(GO:0031337) regulation of homocysteine metabolic process(GO:0050666) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.0 | 0.5 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.3 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.0 | 0.2 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0070377 | regulation of ERK5 cascade(GO:0070376) negative regulation of ERK5 cascade(GO:0070377) |
| 0.0 | 0.3 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.1 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.2 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.1 | GO:0090675 | intermicrovillar adhesion(GO:0090675) |
| 0.0 | 0.3 | GO:0032096 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.0 | 0.2 | GO:0060268 | negative regulation of respiratory burst(GO:0060268) |
| 0.0 | 0.1 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.0 | 0.1 | GO:0009183 | purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 0.0 | 0.1 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.0 | 0.2 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.1 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.0 | 0.1 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.2 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.1 | GO:0048817 | negative regulation of hair follicle maturation(GO:0048817) |
| 0.0 | 0.1 | GO:0097531 | mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.0 | 0.0 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.0 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.2 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.0 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.0 | GO:0003285 | septum secundum development(GO:0003285) |
| 0.0 | 0.1 | GO:1900154 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.0 | 0.1 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.0 | 0.3 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) |
| 0.0 | 0.2 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.0 | 0.1 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.0 | 0.1 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.1 | GO:2000591 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.0 | 0.4 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.0 | 0.5 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.0 | GO:0043257 | laminin-8 complex(GO:0043257) |
| 0.0 | 0.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.3 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.0 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.9 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.2 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.1 | GO:0004397 | histidine ammonia-lyase activity(GO:0004397) |
| 0.0 | 0.2 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.3 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.3 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.2 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.0 | 0.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0008841 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) dihydrofolate synthase activity(GO:0008841) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0004952 | dopamine neurotransmitter receptor activity(GO:0004952) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.0 | 0.1 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.0 | 0.0 | GO:0005019 | platelet-derived growth factor beta-receptor activity(GO:0005019) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.3 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.4 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.2 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 1.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.4 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.2 | ST STAT3 PATHWAY | STAT3 Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |