Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
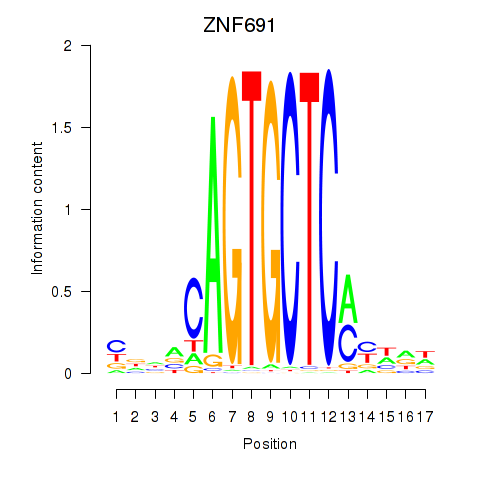
Results for ZNF691
Z-value: 0.51
Transcription factors associated with ZNF691
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF691
|
ENSG00000164011.13 | zinc finger protein 691 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF691 | hg19_v2_chr1_+_43312258_43312310 | -0.38 | 6.2e-01 | Click! |
Activity profile of ZNF691 motif
Sorted Z-values of ZNF691 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_28199047 | 1.09 |
ENST00000373925.1
ENST00000328928.7 ENST00000373927.3 ENST00000427466.1 ENST00000442118.1 ENST00000373921.3 |
THEMIS2
|
thymocyte selection associated family member 2 |
| chr1_+_223354486 | 0.57 |
ENST00000446145.1
|
RP11-239E10.3
|
RP11-239E10.3 |
| chr1_-_935361 | 0.56 |
ENST00000484667.2
|
HES4
|
hes family bHLH transcription factor 4 |
| chr15_-_40398812 | 0.41 |
ENST00000561360.1
|
BMF
|
Bcl2 modifying factor |
| chr10_-_4720301 | 0.36 |
ENST00000449712.1
|
LINC00704
|
long intergenic non-protein coding RNA 704 |
| chr14_+_55033815 | 0.29 |
ENST00000554335.1
|
SAMD4A
|
sterile alpha motif domain containing 4A |
| chr2_+_238475217 | 0.28 |
ENST00000165524.1
|
PRLH
|
prolactin releasing hormone |
| chr3_+_183899770 | 0.28 |
ENST00000442686.1
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit |
| chr19_-_5903714 | 0.25 |
ENST00000586349.1
ENST00000585661.1 ENST00000308961.4 ENST00000592634.1 ENST00000418389.2 ENST00000252675.5 |
AC024592.12
NDUFA11
FUT5
|
Uncharacterized protein NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa fucosyltransferase 5 (alpha (1,3) fucosyltransferase) |
| chr17_+_79495397 | 0.23 |
ENST00000417245.2
ENST00000334850.7 |
FSCN2
|
fascin homolog 2, actin-bundling protein, retinal (Strongylocentrotus purpuratus) |
| chr8_-_123793048 | 0.23 |
ENST00000607710.1
|
RP11-44N11.2
|
RP11-44N11.2 |
| chr13_-_52703187 | 0.22 |
ENST00000355568.4
|
NEK5
|
NIMA-related kinase 5 |
| chr10_-_5638048 | 0.21 |
ENST00000478294.1
|
RP13-463N16.6
|
RP13-463N16.6 |
| chr5_-_39074479 | 0.20 |
ENST00000514735.1
ENST00000296782.5 ENST00000357387.3 |
RICTOR
|
RPTOR independent companion of MTOR, complex 2 |
| chr11_-_133826852 | 0.20 |
ENST00000533871.2
ENST00000321016.8 |
IGSF9B
|
immunoglobulin superfamily, member 9B |
| chr8_+_27168988 | 0.19 |
ENST00000397501.1
ENST00000338238.4 ENST00000544172.1 |
PTK2B
|
protein tyrosine kinase 2 beta |
| chr14_+_55034330 | 0.19 |
ENST00000251091.5
|
SAMD4A
|
sterile alpha motif domain containing 4A |
| chr17_-_36760865 | 0.19 |
ENST00000584266.1
|
SRCIN1
|
SRC kinase signaling inhibitor 1 |
| chr22_-_32058416 | 0.19 |
ENST00000439502.2
|
PISD
|
phosphatidylserine decarboxylase |
| chr10_-_4720333 | 0.18 |
ENST00000430998.2
|
LINC00704
|
long intergenic non-protein coding RNA 704 |
| chr8_+_145294015 | 0.17 |
ENST00000544576.1
|
MROH1
|
maestro heat-like repeat family member 1 |
| chr4_-_120243545 | 0.16 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr1_+_39796810 | 0.16 |
ENST00000289893.4
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr16_+_31225337 | 0.15 |
ENST00000322122.3
|
TRIM72
|
tripartite motif containing 72 |
| chr11_+_102188272 | 0.15 |
ENST00000532808.1
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr5_+_135170331 | 0.14 |
ENST00000425402.1
ENST00000274513.5 ENST00000420621.1 ENST00000433282.2 ENST00000412661.2 |
SLC25A48
|
solute carrier family 25, member 48 |
| chr19_+_50380917 | 0.14 |
ENST00000535102.2
|
TBC1D17
|
TBC1 domain family, member 17 |
| chr1_+_15986364 | 0.14 |
ENST00000345034.1
|
RSC1A1
|
regulatory solute carrier protein, family 1, member 1 |
| chr10_+_4828815 | 0.14 |
ENST00000533295.1
|
AKR1E2
|
aldo-keto reductase family 1, member E2 |
| chr19_+_40877583 | 0.13 |
ENST00000596470.1
|
PLD3
|
phospholipase D family, member 3 |
| chr17_-_80797886 | 0.13 |
ENST00000572562.1
|
ZNF750
|
zinc finger protein 750 |
| chrX_+_107069063 | 0.11 |
ENST00000262843.6
|
MID2
|
midline 2 |
| chr22_-_45608237 | 0.11 |
ENST00000492273.1
|
KIAA0930
|
KIAA0930 |
| chr3_+_177534653 | 0.11 |
ENST00000436078.1
|
RP11-91K9.1
|
RP11-91K9.1 |
| chr15_-_72767490 | 0.11 |
ENST00000565181.1
|
RP11-1007O24.3
|
RP11-1007O24.3 |
| chr9_+_139553306 | 0.11 |
ENST00000371699.1
|
EGFL7
|
EGF-like-domain, multiple 7 |
| chr3_+_44596679 | 0.11 |
ENST00000426540.1
ENST00000431636.1 ENST00000341840.3 ENST00000273320.3 |
ZKSCAN7
|
zinc finger with KRAB and SCAN domains 7 |
| chr10_+_104678032 | 0.10 |
ENST00000369878.4
ENST00000369875.3 |
CNNM2
|
cyclin M2 |
| chr17_+_30771279 | 0.10 |
ENST00000261712.3
ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chrX_+_153672468 | 0.10 |
ENST00000393600.3
|
FAM50A
|
family with sequence similarity 50, member A |
| chr5_-_179107975 | 0.10 |
ENST00000376974.4
|
CBY3
|
chibby homolog 3 (Drosophila) |
| chr3_-_194968576 | 0.09 |
ENST00000429994.1
|
XXYLT1
|
xyloside xylosyltransferase 1 |
| chr8_+_27169138 | 0.09 |
ENST00000522338.1
|
PTK2B
|
protein tyrosine kinase 2 beta |
| chr12_-_109221160 | 0.09 |
ENST00000326470.5
|
SSH1
|
slingshot protein phosphatase 1 |
| chr17_+_36908984 | 0.09 |
ENST00000225426.4
ENST00000579088.1 |
PSMB3
|
proteasome (prosome, macropain) subunit, beta type, 3 |
| chr12_+_27619743 | 0.09 |
ENST00000298876.4
ENST00000416383.1 |
SMCO2
|
single-pass membrane protein with coiled-coil domains 2 |
| chr22_-_32058166 | 0.08 |
ENST00000435900.1
ENST00000336566.4 |
PISD
|
phosphatidylserine decarboxylase |
| chr19_+_50084561 | 0.08 |
ENST00000246794.5
|
PRRG2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr9_+_71944241 | 0.08 |
ENST00000257515.8
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr2_-_110371664 | 0.08 |
ENST00000545389.1
ENST00000423520.1 |
SEPT10
|
septin 10 |
| chr20_-_2644832 | 0.08 |
ENST00000380851.5
ENST00000380843.4 |
IDH3B
|
isocitrate dehydrogenase 3 (NAD+) beta |
| chr20_-_33735070 | 0.07 |
ENST00000374491.3
ENST00000542871.1 ENST00000374492.3 |
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr16_-_1821496 | 0.07 |
ENST00000564628.1
ENST00000563498.1 |
NME3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr19_+_55043977 | 0.07 |
ENST00000335056.3
|
KIR3DX1
|
killer cell immunoglobulin-like receptor, three domains, X1 |
| chr6_+_31553978 | 0.07 |
ENST00000376096.1
ENST00000376099.1 ENST00000376110.3 |
LST1
|
leukocyte specific transcript 1 |
| chr11_+_44748361 | 0.07 |
ENST00000533202.1
ENST00000533080.1 ENST00000520358.2 ENST00000520999.2 |
TSPAN18
|
tetraspanin 18 |
| chr2_+_202098166 | 0.07 |
ENST00000392263.2
ENST00000264274.9 ENST00000392259.2 ENST00000392266.3 ENST00000432109.2 ENST00000264275.5 |
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr3_-_49907323 | 0.07 |
ENST00000296471.7
ENST00000488336.1 ENST00000467248.1 ENST00000466940.1 ENST00000463537.1 ENST00000480398.2 |
CAMKV
|
CaM kinase-like vesicle-associated |
| chr3_-_167452614 | 0.07 |
ENST00000392750.2
ENST00000464360.1 ENST00000492139.1 ENST00000471885.1 ENST00000470131.1 |
PDCD10
|
programmed cell death 10 |
| chr21_-_37852359 | 0.07 |
ENST00000399137.1
ENST00000399135.1 |
CLDN14
|
claudin 14 |
| chr13_-_52027134 | 0.07 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr16_+_78133293 | 0.06 |
ENST00000566780.1
|
WWOX
|
WW domain containing oxidoreductase |
| chr17_-_61777459 | 0.06 |
ENST00000578993.1
ENST00000583211.1 ENST00000259006.3 |
LIMD2
|
LIM domain containing 2 |
| chr3_-_157251383 | 0.06 |
ENST00000487753.1
ENST00000489602.1 ENST00000461299.1 ENST00000479987.1 |
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr19_-_893200 | 0.06 |
ENST00000269814.4
ENST00000395808.3 ENST00000312090.6 ENST00000325464.1 |
MED16
|
mediator complex subunit 16 |
| chr5_+_53686658 | 0.06 |
ENST00000512618.1
|
LINC01033
|
long intergenic non-protein coding RNA 1033 |
| chrX_-_153775426 | 0.06 |
ENST00000393562.2
|
G6PD
|
glucose-6-phosphate dehydrogenase |
| chrX_+_102469997 | 0.06 |
ENST00000372695.5
ENST00000372691.3 |
BEX4
|
brain expressed, X-linked 4 |
| chr10_-_61122220 | 0.06 |
ENST00000422313.2
ENST00000435852.2 ENST00000442566.3 ENST00000373868.2 ENST00000277705.6 ENST00000373867.3 ENST00000419214.2 |
FAM13C
|
family with sequence similarity 13, member C |
| chr19_-_15560730 | 0.06 |
ENST00000389282.4
ENST00000263381.7 |
WIZ
|
widely interspaced zinc finger motifs |
| chr10_-_91011548 | 0.05 |
ENST00000456827.1
|
LIPA
|
lipase A, lysosomal acid, cholesterol esterase |
| chr8_-_28347737 | 0.05 |
ENST00000517673.1
ENST00000518734.1 ENST00000346498.2 ENST00000380254.2 |
FBXO16
|
F-box protein 16 |
| chr2_+_202098203 | 0.05 |
ENST00000450491.1
ENST00000440732.1 ENST00000392258.3 |
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr8_+_32579341 | 0.05 |
ENST00000519240.1
ENST00000539990.1 |
NRG1
|
neuregulin 1 |
| chr1_-_155232221 | 0.05 |
ENST00000355379.3
|
SCAMP3
|
secretory carrier membrane protein 3 |
| chr1_-_233431458 | 0.05 |
ENST00000258229.9
ENST00000430153.1 |
PCNXL2
|
pecanex-like 2 (Drosophila) |
| chr14_-_106725723 | 0.05 |
ENST00000390609.2
|
IGHV3-23
|
immunoglobulin heavy variable 3-23 |
| chr3_-_167452262 | 0.05 |
ENST00000487947.2
|
PDCD10
|
programmed cell death 10 |
| chr2_+_202125219 | 0.04 |
ENST00000323492.7
|
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chrX_+_16188506 | 0.04 |
ENST00000329538.5
|
MAGEB17
|
melanoma antigen family B, 17 |
| chr7_-_143059780 | 0.04 |
ENST00000409578.1
ENST00000409346.1 |
FAM131B
|
family with sequence similarity 131, member B |
| chr6_+_31553901 | 0.04 |
ENST00000418507.2
ENST00000438075.2 ENST00000376100.3 ENST00000376111.4 |
LST1
|
leukocyte specific transcript 1 |
| chr2_-_85788652 | 0.04 |
ENST00000430215.3
|
GGCX
|
gamma-glutamyl carboxylase |
| chr11_+_121461097 | 0.04 |
ENST00000527934.1
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr12_-_10605929 | 0.04 |
ENST00000347831.5
ENST00000359151.3 |
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr4_-_40516560 | 0.03 |
ENST00000513473.1
|
RBM47
|
RNA binding motif protein 47 |
| chr16_+_810728 | 0.03 |
ENST00000563941.1
ENST00000545450.2 ENST00000566549.1 |
MSLN
|
mesothelin |
| chr8_+_32579321 | 0.03 |
ENST00000522402.1
|
NRG1
|
neuregulin 1 |
| chr5_-_42887494 | 0.03 |
ENST00000514218.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr22_-_43539346 | 0.03 |
ENST00000327555.5
ENST00000290429.6 |
MCAT
|
malonyl CoA:ACP acyltransferase (mitochondrial) |
| chr17_-_3417062 | 0.03 |
ENST00000570318.1
ENST00000541913.1 |
SPATA22
|
spermatogenesis associated 22 |
| chr17_+_73606766 | 0.03 |
ENST00000578462.1
|
MYO15B
|
myosin XVB pseudogene |
| chrX_+_107068959 | 0.02 |
ENST00000451923.1
|
MID2
|
midline 2 |
| chr11_-_71750865 | 0.02 |
ENST00000544129.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr17_+_80416482 | 0.02 |
ENST00000309794.11
ENST00000345415.7 ENST00000457415.3 ENST00000584411.1 ENST00000412079.2 ENST00000577432.1 |
NARF
|
nuclear prelamin A recognition factor |
| chr2_-_85895295 | 0.02 |
ENST00000428225.1
ENST00000519937.2 |
SFTPB
|
surfactant protein B |
| chr9_+_116267536 | 0.02 |
ENST00000374136.1
|
RGS3
|
regulator of G-protein signaling 3 |
| chr1_-_148202536 | 0.02 |
ENST00000544708.1
|
PPIAL4D
|
peptidylprolyl isomerase A (cyclophilin A)-like 4D |
| chr12_+_19389814 | 0.02 |
ENST00000536974.1
|
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr2_+_68961934 | 0.02 |
ENST00000409202.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr18_+_11751466 | 0.02 |
ENST00000535121.1
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr16_+_23765948 | 0.02 |
ENST00000300113.2
|
CHP2
|
calcineurin-like EF-hand protein 2 |
| chr1_+_33938236 | 0.02 |
ENST00000361328.3
ENST00000373413.2 |
ZSCAN20
|
zinc finger and SCAN domain containing 20 |
| chr7_+_7811992 | 0.02 |
ENST00000406829.1
|
RPA3-AS1
|
RPA3 antisense RNA 1 |
| chr20_-_42816206 | 0.01 |
ENST00000372980.3
|
JPH2
|
junctophilin 2 |
| chr6_-_27279949 | 0.01 |
ENST00000444565.1
ENST00000377451.2 |
POM121L2
|
POM121 transmembrane nucleoporin-like 2 |
| chr12_-_51718436 | 0.01 |
ENST00000544402.1
|
BIN2
|
bridging integrator 2 |
| chr16_+_85687999 | 0.01 |
ENST00000412692.1
|
GSE1
|
Gse1 coiled-coil protein |
| chr17_+_80416050 | 0.01 |
ENST00000579198.1
ENST00000390006.4 ENST00000580296.1 |
NARF
|
nuclear prelamin A recognition factor |
| chr2_+_68961905 | 0.01 |
ENST00000295381.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr1_-_38455650 | 0.01 |
ENST00000448721.2
|
SF3A3
|
splicing factor 3a, subunit 3, 60kDa |
| chr17_+_37026106 | 0.01 |
ENST00000318008.6
|
LASP1
|
LIM and SH3 protein 1 |
| chrX_+_153775821 | 0.01 |
ENST00000263518.6
ENST00000470142.1 ENST00000393549.2 ENST00000455588.2 ENST00000369602.3 |
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr4_+_48492269 | 0.01 |
ENST00000327939.4
|
ZAR1
|
zygote arrest 1 |
| chr4_-_16077741 | 0.01 |
ENST00000447510.2
ENST00000540805.1 ENST00000539194.1 |
PROM1
|
prominin 1 |
| chr8_+_144821557 | 0.01 |
ENST00000534398.1
|
FAM83H-AS1
|
FAM83H antisense RNA 1 (head to head) |
| chr6_+_31021225 | 0.01 |
ENST00000565192.1
ENST00000562344.1 |
HCG22
|
HLA complex group 22 |
| chr3_+_37493610 | 0.01 |
ENST00000264741.5
|
ITGA9
|
integrin, alpha 9 |
| chr19_+_40873617 | 0.01 |
ENST00000599353.1
|
PLD3
|
phospholipase D family, member 3 |
| chr12_+_120502441 | 0.01 |
ENST00000446727.2
|
CCDC64
|
coiled-coil domain containing 64 |
| chr14_+_60712463 | 0.01 |
ENST00000325642.3
ENST00000529574.1 |
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr16_-_11492366 | 0.01 |
ENST00000595360.1
|
CTD-3088G3.8
|
Protein LOC388210 |
| chr20_-_2489542 | 0.00 |
ENST00000421216.1
ENST00000381253.1 |
ZNF343
|
zinc finger protein 343 |
| chr17_+_20978854 | 0.00 |
ENST00000456235.1
|
AC087393.1
|
AC087393.1 |
| chr17_+_37026284 | 0.00 |
ENST00000433206.2
ENST00000435347.3 |
LASP1
|
LIM and SH3 protein 1 |
| chr15_+_84904525 | 0.00 |
ENST00000510439.2
|
GOLGA6L4
|
golgin A6 family-like 4 |
| chr4_+_657485 | 0.00 |
ENST00000471824.2
|
PDE6B
|
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr12_-_51717948 | 0.00 |
ENST00000267012.4
|
BIN2
|
bridging integrator 2 |
| chr9_+_138453595 | 0.00 |
ENST00000479141.1
ENST00000371766.2 ENST00000277508.5 ENST00000433563.1 |
PAEP
|
progestagen-associated endometrial protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF691
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.3 | GO:2000537 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.2 | GO:0052053 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 0.1 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.3 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.1 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.4 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0036511 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.2 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.0 | GO:1902997 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) regulation of neurofibrillary tangle assembly(GO:1902996) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.3 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.0 | 0.3 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |