Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
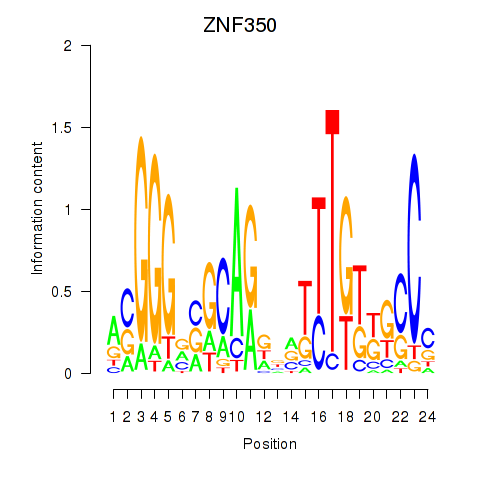
Results for ZNF350
Z-value: 0.88
Transcription factors associated with ZNF350
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF350
|
ENSG00000256683.2 | zinc finger protein 350 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF350 | hg19_v2_chr19_-_52489923_52490113 | -0.84 | 1.6e-01 | Click! |
Activity profile of ZNF350 motif
Sorted Z-values of ZNF350 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_327537 | 1.83 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr19_+_35532612 | 1.07 |
ENST00000600390.1
ENST00000597419.1 |
HPN
|
hepsin |
| chr11_+_118826999 | 1.06 |
ENST00000264031.2
|
UPK2
|
uroplakin 2 |
| chr12_+_113376249 | 1.01 |
ENST00000551007.1
ENST00000548514.1 |
OAS3
|
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr16_-_74734742 | 0.84 |
ENST00000308807.7
ENST00000573267.1 |
MLKL
|
mixed lineage kinase domain-like |
| chr12_+_113376157 | 0.70 |
ENST00000228928.7
|
OAS3
|
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr11_+_64323156 | 0.65 |
ENST00000377585.3
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr17_-_39674668 | 0.65 |
ENST00000393981.3
|
KRT15
|
keratin 15 |
| chr2_-_106810783 | 0.62 |
ENST00000283148.7
|
UXS1
|
UDP-glucuronate decarboxylase 1 |
| chr2_-_106810742 | 0.55 |
ENST00000409501.3
ENST00000428048.2 ENST00000441952.1 ENST00000457835.1 ENST00000540130.1 |
UXS1
|
UDP-glucuronate decarboxylase 1 |
| chr11_-_64511789 | 0.52 |
ENST00000419843.1
ENST00000394430.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr3_-_128212016 | 0.51 |
ENST00000498200.1
ENST00000341105.2 |
GATA2
|
GATA binding protein 2 |
| chr17_+_68165657 | 0.50 |
ENST00000243457.3
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr2_+_233497931 | 0.49 |
ENST00000264059.3
|
EFHD1
|
EF-hand domain family, member D1 |
| chr11_-_45687128 | 0.48 |
ENST00000308064.2
|
CHST1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 |
| chr11_-_64511575 | 0.46 |
ENST00000431822.1
ENST00000377486.3 ENST00000394432.3 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr22_-_50964558 | 0.44 |
ENST00000535425.1
ENST00000439934.1 |
SCO2
|
SCO2 cytochrome c oxidase assembly protein |
| chr4_-_99578789 | 0.44 |
ENST00000511651.1
ENST00000505184.1 |
TSPAN5
|
tetraspanin 5 |
| chr1_+_228395755 | 0.42 |
ENST00000284548.11
ENST00000570156.2 ENST00000422127.1 ENST00000366707.4 ENST00000366709.4 |
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr11_+_64323428 | 0.42 |
ENST00000377581.3
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr11_+_48002279 | 0.42 |
ENST00000534219.1
ENST00000527952.1 |
PTPRJ
|
protein tyrosine phosphatase, receptor type, J |
| chr9_-_117267717 | 0.41 |
ENST00000374057.3
|
DFNB31
|
deafness, autosomal recessive 31 |
| chr13_-_99738867 | 0.37 |
ENST00000427887.1
|
DOCK9
|
dedicator of cytokinesis 9 |
| chr12_-_49351148 | 0.37 |
ENST00000398092.4
ENST00000539611.1 |
RP11-302B13.5
ARF3
|
ADP-ribosylation factor 3 ADP-ribosylation factor 3 |
| chr3_-_118753792 | 0.35 |
ENST00000480431.1
|
IGSF11
|
immunoglobulin superfamily, member 11 |
| chr19_+_39903185 | 0.35 |
ENST00000409794.3
|
PLEKHG2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr2_-_70780770 | 0.33 |
ENST00000444975.1
ENST00000445399.1 ENST00000418333.2 |
TGFA
|
transforming growth factor, alpha |
| chr10_-_94003003 | 0.32 |
ENST00000412050.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr11_-_64647144 | 0.30 |
ENST00000359393.2
ENST00000433803.1 ENST00000411683.1 |
EHD1
|
EH-domain containing 1 |
| chr1_-_1822495 | 0.29 |
ENST00000378609.4
|
GNB1
|
guanine nucleotide binding protein (G protein), beta polypeptide 1 |
| chr11_+_48002076 | 0.29 |
ENST00000418331.2
ENST00000440289.2 |
PTPRJ
|
protein tyrosine phosphatase, receptor type, J |
| chr17_-_18950950 | 0.27 |
ENST00000284154.5
|
GRAP
|
GRB2-related adaptor protein |
| chr16_+_69600209 | 0.27 |
ENST00000566899.1
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive |
| chrX_+_56259316 | 0.27 |
ENST00000468660.1
|
KLF8
|
Kruppel-like factor 8 |
| chr8_-_119124045 | 0.27 |
ENST00000378204.2
|
EXT1
|
exostosin glycosyltransferase 1 |
| chr5_+_42423872 | 0.26 |
ENST00000230882.4
ENST00000357703.3 |
GHR
|
growth hormone receptor |
| chr17_-_18950310 | 0.26 |
ENST00000573099.1
|
GRAP
|
GRB2-related adaptor protein |
| chr12_-_53228079 | 0.26 |
ENST00000330553.5
|
KRT79
|
keratin 79 |
| chr17_+_19030782 | 0.25 |
ENST00000344415.4
ENST00000577213.1 |
GRAPL
|
GRB2-related adaptor protein-like |
| chr14_+_73603126 | 0.23 |
ENST00000557356.1
ENST00000556864.1 ENST00000556533.1 ENST00000556951.1 ENST00000557293.1 ENST00000553719.1 ENST00000553599.1 ENST00000556011.1 ENST00000394157.3 ENST00000357710.4 ENST00000324501.5 ENST00000560005.2 ENST00000555254.1 ENST00000261970.3 ENST00000344094.3 ENST00000554131.1 ENST00000557037.1 |
PSEN1
|
presenilin 1 |
| chr16_+_67282853 | 0.22 |
ENST00000299798.11
|
SLC9A5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr16_+_14802801 | 0.22 |
ENST00000526520.1
ENST00000531598.2 |
NPIPA3
|
nuclear pore complex interacting protein family, member A3 |
| chr17_-_6554877 | 0.22 |
ENST00000225728.3
ENST00000575197.1 |
MED31
|
mediator complex subunit 31 |
| chr4_-_76008706 | 0.21 |
ENST00000562355.1
ENST00000563602.1 |
RP11-44F21.5
|
RP11-44F21.5 |
| chr9_-_92051354 | 0.20 |
ENST00000418828.1
|
SEMA4D
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
| chr20_+_62367989 | 0.20 |
ENST00000309546.3
|
LIME1
|
Lck interacting transmembrane adaptor 1 |
| chr17_+_46985823 | 0.20 |
ENST00000508468.2
|
UBE2Z
|
ubiquitin-conjugating enzyme E2Z |
| chr17_+_7487146 | 0.19 |
ENST00000396501.4
ENST00000584378.1 ENST00000423172.2 ENST00000579445.1 ENST00000585217.1 ENST00000581380.1 |
MPDU1
|
mannose-P-dolichol utilization defect 1 |
| chr19_+_2977444 | 0.19 |
ENST00000246112.4
ENST00000453329.1 ENST00000482627.1 ENST00000452088.1 |
TLE6
|
transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) |
| chr19_+_5681153 | 0.19 |
ENST00000579559.1
ENST00000577222.1 |
HSD11B1L
RPL36
|
hydroxysteroid (11-beta) dehydrogenase 1-like ribosomal protein L36 |
| chr14_+_96671016 | 0.19 |
ENST00000542454.2
ENST00000554311.1 ENST00000306005.3 ENST00000539359.1 ENST00000553811.1 |
BDKRB2
RP11-404P21.8
|
bradykinin receptor B2 Uncharacterized protein |
| chr11_+_8932828 | 0.18 |
ENST00000530281.1
ENST00000396648.2 ENST00000534147.1 ENST00000529942.1 |
AKIP1
|
A kinase (PRKA) interacting protein 1 |
| chr17_+_6554971 | 0.17 |
ENST00000391428.2
|
C17orf100
|
chromosome 17 open reading frame 100 |
| chr16_+_31225337 | 0.17 |
ENST00000322122.3
|
TRIM72
|
tripartite motif containing 72 |
| chr11_-_414948 | 0.16 |
ENST00000530494.1
ENST00000528209.1 ENST00000431843.2 ENST00000528058.1 |
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr2_+_120301997 | 0.16 |
ENST00000602047.1
|
PCDP1
|
Primary ciliary dyskinesia protein 1 |
| chr11_+_8932715 | 0.15 |
ENST00000529876.1
ENST00000525005.1 ENST00000524577.1 ENST00000534506.1 |
AKIP1
|
A kinase (PRKA) interacting protein 1 |
| chr2_+_235860690 | 0.14 |
ENST00000416021.1
|
SH3BP4
|
SH3-domain binding protein 4 |
| chr20_+_19738300 | 0.14 |
ENST00000432334.1
|
RP1-122P22.2
|
RP1-122P22.2 |
| chr9_-_92051391 | 0.13 |
ENST00000420681.2
|
SEMA4D
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
| chr11_+_65779283 | 0.13 |
ENST00000312134.2
|
CST6
|
cystatin E/M |
| chr11_+_10326918 | 0.13 |
ENST00000528544.1
|
ADM
|
adrenomedullin |
| chr16_-_74734672 | 0.13 |
ENST00000306247.7
ENST00000575686.1 |
MLKL
|
mixed lineage kinase domain-like |
| chr19_-_5680499 | 0.13 |
ENST00000587589.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr2_+_120302041 | 0.12 |
ENST00000442513.3
ENST00000413369.3 |
PCDP1
|
Primary ciliary dyskinesia protein 1 |
| chr19_-_5680891 | 0.12 |
ENST00000309324.4
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr2_+_132286754 | 0.12 |
ENST00000434330.1
|
CCDC74A
|
coiled-coil domain containing 74A |
| chr19_+_35645618 | 0.12 |
ENST00000392218.2
ENST00000543307.1 ENST00000392219.2 ENST00000541435.2 ENST00000590686.1 ENST00000342879.3 ENST00000588699.1 |
FXYD5
|
FXYD domain containing ion transport regulator 5 |
| chr1_-_33647267 | 0.11 |
ENST00000291416.5
|
TRIM62
|
tripartite motif containing 62 |
| chr1_+_63788730 | 0.11 |
ENST00000371116.2
|
FOXD3
|
forkhead box D3 |
| chr2_-_21266816 | 0.11 |
ENST00000399256.4
|
APOB
|
apolipoprotein B |
| chr2_+_235860616 | 0.10 |
ENST00000392011.2
|
SH3BP4
|
SH3-domain binding protein 4 |
| chr19_+_57050317 | 0.10 |
ENST00000301318.3
ENST00000591844.1 |
ZFP28
|
ZFP28 zinc finger protein |
| chr11_+_7041677 | 0.10 |
ENST00000299481.4
|
NLRP14
|
NLR family, pyrin domain containing 14 |
| chr19_+_49866851 | 0.10 |
ENST00000221498.2
ENST00000596402.1 |
DKKL1
|
dickkopf-like 1 |
| chr11_+_57531292 | 0.09 |
ENST00000524579.1
|
CTNND1
|
catenin (cadherin-associated protein), delta 1 |
| chr16_+_71879861 | 0.09 |
ENST00000427980.2
ENST00000568581.1 |
ATXN1L
IST1
|
ataxin 1-like increased sodium tolerance 1 homolog (yeast) |
| chr2_-_9143786 | 0.09 |
ENST00000462696.1
ENST00000305997.3 |
MBOAT2
|
membrane bound O-acyltransferase domain containing 2 |
| chr1_+_22970119 | 0.09 |
ENST00000374640.4
ENST00000374639.3 ENST00000374637.1 |
C1QC
|
complement component 1, q subcomponent, C chain |
| chr2_-_241759622 | 0.09 |
ENST00000320389.7
ENST00000498729.2 |
KIF1A
|
kinesin family member 1A |
| chr8_-_100905363 | 0.09 |
ENST00000524245.1
|
COX6C
|
cytochrome c oxidase subunit VIc |
| chr19_+_35645817 | 0.08 |
ENST00000423817.3
|
FXYD5
|
FXYD domain containing ion transport regulator 5 |
| chr11_+_8932654 | 0.08 |
ENST00000299576.5
ENST00000309377.4 ENST00000309357.4 |
AKIP1
|
A kinase (PRKA) interacting protein 1 |
| chr2_-_219157250 | 0.08 |
ENST00000434015.2
ENST00000444183.1 ENST00000420341.1 ENST00000453281.1 ENST00000258412.3 ENST00000440422.1 |
TMBIM1
|
transmembrane BAX inhibitor motif containing 1 |
| chr2_+_177025619 | 0.08 |
ENST00000410016.1
|
HOXD3
|
homeobox D3 |
| chr7_+_2559399 | 0.08 |
ENST00000222725.5
ENST00000359574.3 |
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr17_-_39968855 | 0.07 |
ENST00000355468.3
ENST00000590496.1 |
LEPREL4
|
leprecan-like 4 |
| chr2_-_219157219 | 0.07 |
ENST00000445635.1
ENST00000413976.1 |
TMBIM1
|
transmembrane BAX inhibitor motif containing 1 |
| chr17_-_8093471 | 0.07 |
ENST00000389017.4
|
C17orf59
|
chromosome 17 open reading frame 59 |
| chr10_-_15762124 | 0.07 |
ENST00000378076.3
|
ITGA8
|
integrin, alpha 8 |
| chr1_-_155948218 | 0.07 |
ENST00000313667.4
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr1_+_156611960 | 0.07 |
ENST00000361588.5
|
BCAN
|
brevican |
| chr4_-_5991545 | 0.07 |
ENST00000531445.1
|
C4orf50
|
chromosome 4 open reading frame 50 |
| chr11_-_63381823 | 0.06 |
ENST00000323646.5
|
PLA2G16
|
phospholipase A2, group XVI |
| chr15_-_43029252 | 0.06 |
ENST00000563260.1
ENST00000356231.3 |
CDAN1
|
codanin 1 |
| chr19_+_5681011 | 0.05 |
ENST00000581893.1
ENST00000411793.2 ENST00000301382.4 ENST00000581773.1 ENST00000423665.2 ENST00000583928.1 ENST00000342970.2 ENST00000422535.2 ENST00000581521.1 ENST00000339423.2 |
HSD11B1L
|
hydroxysteroid (11-beta) dehydrogenase 1-like |
| chr3_+_100328433 | 0.05 |
ENST00000273352.3
|
GPR128
|
G protein-coupled receptor 128 |
| chr13_-_37633567 | 0.05 |
ENST00000464744.1
|
SUPT20H
|
suppressor of Ty 20 homolog (S. cerevisiae) |
| chr10_-_97175444 | 0.05 |
ENST00000486141.2
|
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr2_+_120302055 | 0.05 |
ENST00000598644.1
|
PCDP1
|
Primary ciliary dyskinesia protein 1 |
| chr12_+_109569155 | 0.05 |
ENST00000539864.1
|
ACACB
|
acetyl-CoA carboxylase beta |
| chrX_-_47479246 | 0.05 |
ENST00000295987.7
ENST00000340666.4 |
SYN1
|
synapsin I |
| chr11_-_68671264 | 0.05 |
ENST00000362034.2
|
MRPL21
|
mitochondrial ribosomal protein L21 |
| chr6_+_36973406 | 0.04 |
ENST00000274963.8
|
FGD2
|
FYVE, RhoGEF and PH domain containing 2 |
| chr1_-_51425902 | 0.04 |
ENST00000396153.2
|
FAF1
|
Fas (TNFRSF6) associated factor 1 |
| chr1_+_156611900 | 0.04 |
ENST00000457777.2
ENST00000424639.1 |
BCAN
|
brevican |
| chr1_+_204797749 | 0.04 |
ENST00000367172.4
ENST00000367171.4 ENST00000367170.4 ENST00000338515.6 ENST00000339876.6 ENST00000338586.6 ENST00000539706.1 ENST00000360049.4 ENST00000367169.4 ENST00000446412.1 ENST00000403080.1 |
NFASC
|
neurofascin |
| chr3_-_113775328 | 0.04 |
ENST00000483766.1
ENST00000545063.1 ENST00000491000.1 ENST00000295878.3 |
KIAA1407
|
KIAA1407 |
| chr19_+_49866331 | 0.04 |
ENST00000597873.1
|
DKKL1
|
dickkopf-like 1 |
| chr20_-_35374456 | 0.04 |
ENST00000373803.2
ENST00000359675.2 ENST00000540765.1 ENST00000349004.1 |
NDRG3
|
NDRG family member 3 |
| chr20_+_44035200 | 0.04 |
ENST00000372717.1
ENST00000360981.4 |
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr12_-_109219937 | 0.03 |
ENST00000546697.1
|
SSH1
|
slingshot protein phosphatase 1 |
| chr18_+_77155856 | 0.03 |
ENST00000253506.5
ENST00000591814.1 |
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
| chr17_-_74449252 | 0.03 |
ENST00000319380.7
|
UBE2O
|
ubiquitin-conjugating enzyme E2O |
| chr3_+_14219858 | 0.03 |
ENST00000306024.3
|
LSM3
|
LSM3 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr14_+_96829814 | 0.03 |
ENST00000555181.1
ENST00000553699.1 ENST00000554182.1 |
GSKIP
|
GSK3B interacting protein |
| chr14_-_94254821 | 0.03 |
ENST00000393140.1
|
PRIMA1
|
proline rich membrane anchor 1 |
| chr18_-_5892103 | 0.03 |
ENST00000383490.2
|
TMEM200C
|
transmembrane protein 200C |
| chr4_+_108910870 | 0.03 |
ENST00000403312.1
ENST00000603302.1 ENST00000309522.3 |
HADH
|
hydroxyacyl-CoA dehydrogenase |
| chr12_+_54519842 | 0.02 |
ENST00000508564.1
|
RP11-834C11.4
|
RP11-834C11.4 |
| chr1_+_173446405 | 0.02 |
ENST00000340385.5
|
PRDX6
|
peroxiredoxin 6 |
| chr3_+_140947563 | 0.02 |
ENST00000505013.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr3_+_113775594 | 0.01 |
ENST00000479882.1
ENST00000493014.1 |
QTRTD1
|
queuine tRNA-ribosyltransferase domain containing 1 |
| chr1_-_247094628 | 0.01 |
ENST00000366508.1
ENST00000326225.3 ENST00000391829.2 |
AHCTF1
|
AT hook containing transcription factor 1 |
| chrX_-_17878827 | 0.01 |
ENST00000360011.1
|
RAI2
|
retinoic acid induced 2 |
| chr18_+_77155942 | 0.01 |
ENST00000397790.2
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
| chr3_+_50229037 | 0.01 |
ENST00000232461.3
ENST00000433068.1 |
GNAT1
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 1 |
| chr17_-_73511504 | 0.01 |
ENST00000581870.1
|
CASKIN2
|
CASK interacting protein 2 |
| chr11_-_68671244 | 0.01 |
ENST00000567045.1
ENST00000450904.2 |
MRPL21
|
mitochondrial ribosomal protein L21 |
| chr15_+_91445448 | 0.01 |
ENST00000558290.1
ENST00000558853.1 ENST00000559999.1 |
MAN2A2
|
mannosidase, alpha, class 2A, member 2 |
| chr11_-_63381925 | 0.01 |
ENST00000415826.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr19_+_42829702 | 0.01 |
ENST00000334370.4
|
MEGF8
|
multiple EGF-like-domains 8 |
| chr2_-_70780572 | 0.00 |
ENST00000450929.1
|
TGFA
|
transforming growth factor, alpha |
| chr11_+_208815 | 0.00 |
ENST00000530889.1
|
RIC8A
|
RIC8 guanine nucleotide exchange factor A |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF350
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0033320 | UDP-D-xylose metabolic process(GO:0033319) UDP-D-xylose biosynthetic process(GO:0033320) |
| 0.4 | 1.1 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.2 | 0.5 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.1 | 1.7 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.1 | 0.5 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.3 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 | 0.4 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.1 | 1.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.2 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.1 | 1.8 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 0.3 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 0.3 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.7 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.2 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.0 | 0.3 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.2 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.2 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.1 | GO:2000504 | negative regulation of Fas signaling pathway(GO:1902045) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.3 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.1 | GO:0097647 | dimeric G-protein coupled receptor signaling pathway(GO:0038042) calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.5 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.3 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 0.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.6 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.4 | GO:0006206 | pyrimidine nucleobase metabolic process(GO:0006206) |
| 0.0 | 0.3 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.0 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.1 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.2 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.2 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.0 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.1 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 0.0 | 0.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0048040 | UDP-glucuronate decarboxylase activity(GO:0048040) |
| 0.1 | 1.7 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 1.1 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.7 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.2 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.5 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 0.3 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 0.3 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.4 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 1.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 1.0 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 0.5 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.5 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.9 | PID RAS PATHWAY | Regulation of Ras family activation |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 3.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |