Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
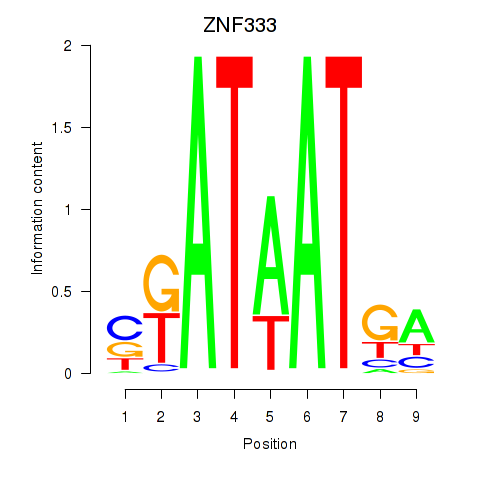
Results for ZNF333
Z-value: 0.57
Transcription factors associated with ZNF333
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF333
|
ENSG00000160961.7 | zinc finger protein 333 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF333 | hg19_v2_chr19_+_14800711_14800917 | -0.71 | 2.9e-01 | Click! |
Activity profile of ZNF333 motif
Sorted Z-values of ZNF333 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_139926236 | 0.54 |
ENST00000303045.6
ENST00000435777.1 |
COL22A1
|
collagen, type XXII, alpha 1 |
| chr5_-_27038683 | 0.53 |
ENST00000511822.1
ENST00000231021.4 |
CDH9
|
cadherin 9, type 2 (T1-cadherin) |
| chr7_+_37723336 | 0.52 |
ENST00000450180.1
|
GPR141
|
G protein-coupled receptor 141 |
| chrX_-_21776281 | 0.44 |
ENST00000379494.3
|
SMPX
|
small muscle protein, X-linked |
| chr22_-_30642728 | 0.43 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr6_+_90192974 | 0.40 |
ENST00000520458.1
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr12_+_112563303 | 0.39 |
ENST00000412615.2
|
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chr12_+_112563335 | 0.36 |
ENST00000549358.1
ENST00000257604.5 ENST00000548092.1 ENST00000552896.1 |
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chr19_+_35741466 | 0.34 |
ENST00000599658.1
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr5_-_78281775 | 0.30 |
ENST00000396151.3
ENST00000565165.1 |
ARSB
|
arylsulfatase B |
| chr17_-_71223839 | 0.30 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr15_+_99433570 | 0.29 |
ENST00000558898.1
|
IGF1R
|
insulin-like growth factor 1 receptor |
| chr5_-_147162078 | 0.27 |
ENST00000507386.1
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2 |
| chr2_+_233527443 | 0.26 |
ENST00000410095.1
|
EFHD1
|
EF-hand domain family, member D1 |
| chr4_+_110749143 | 0.25 |
ENST00000317735.4
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr3_+_173116225 | 0.25 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr10_-_128210005 | 0.25 |
ENST00000284694.7
ENST00000454341.1 ENST00000432642.1 ENST00000392694.1 |
C10orf90
|
chromosome 10 open reading frame 90 |
| chr1_+_225600404 | 0.23 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr5_+_169011033 | 0.23 |
ENST00000513795.1
|
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr1_+_44514040 | 0.23 |
ENST00000431800.1
ENST00000437643.1 |
RP5-1198O20.4
|
RP5-1198O20.4 |
| chr17_-_46690839 | 0.22 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr2_-_127977654 | 0.21 |
ENST00000409327.1
|
CYP27C1
|
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr10_-_4720333 | 0.21 |
ENST00000430998.2
|
LINC00704
|
long intergenic non-protein coding RNA 704 |
| chr3_-_126327398 | 0.19 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr6_+_17110726 | 0.19 |
ENST00000354384.5
|
STMND1
|
stathmin domain containing 1 |
| chr3_-_156878482 | 0.19 |
ENST00000295925.4
|
CCNL1
|
cyclin L1 |
| chr1_+_74701062 | 0.19 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr2_-_175712479 | 0.18 |
ENST00000443238.1
|
CHN1
|
chimerin 1 |
| chr6_-_46620522 | 0.18 |
ENST00000275016.2
|
CYP39A1
|
cytochrome P450, family 39, subfamily A, polypeptide 1 |
| chr12_-_71031220 | 0.18 |
ENST00000334414.6
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr2_-_228497888 | 0.18 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chr21_-_46964306 | 0.17 |
ENST00000443742.1
ENST00000528477.1 ENST00000567670.1 |
SLC19A1
|
solute carrier family 19 (folate transporter), member 1 |
| chr5_-_78281603 | 0.16 |
ENST00000264914.4
|
ARSB
|
arylsulfatase B |
| chrX_-_49121165 | 0.16 |
ENST00000376207.4
ENST00000376199.2 |
FOXP3
|
forkhead box P3 |
| chr9_-_19786926 | 0.15 |
ENST00000341998.2
ENST00000286344.3 |
SLC24A2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 |
| chr10_-_23633720 | 0.15 |
ENST00000323327.4
|
C10orf67
|
chromosome 10 open reading frame 67 |
| chr13_-_46716969 | 0.15 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr8_-_11725549 | 0.14 |
ENST00000505496.2
ENST00000534636.1 ENST00000524500.1 ENST00000345125.3 ENST00000453527.2 ENST00000527215.2 ENST00000532392.1 ENST00000533455.1 ENST00000534510.1 ENST00000530640.2 ENST00000531089.1 ENST00000532656.2 ENST00000531502.1 ENST00000434271.1 ENST00000353047.6 |
CTSB
|
cathepsin B |
| chr13_-_30160925 | 0.14 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr5_+_140800638 | 0.14 |
ENST00000398587.2
ENST00000518882.1 |
PCDHGA11
|
protocadherin gamma subfamily A, 11 |
| chr1_-_36789755 | 0.14 |
ENST00000270824.1
|
EVA1B
|
eva-1 homolog B (C. elegans) |
| chr5_-_54988559 | 0.13 |
ENST00000502247.1
|
SLC38A9
|
solute carrier family 38, member 9 |
| chr20_+_1099233 | 0.13 |
ENST00000246015.4
ENST00000335877.6 ENST00000438768.2 |
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
| chr3_+_38323785 | 0.13 |
ENST00000466887.1
ENST00000448498.1 |
SLC22A14
|
solute carrier family 22, member 14 |
| chr7_-_78400598 | 0.12 |
ENST00000536571.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr1_+_11994715 | 0.12 |
ENST00000449038.1
ENST00000376369.3 ENST00000429000.2 ENST00000196061.4 |
PLOD1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr2_+_217363559 | 0.12 |
ENST00000600880.1
ENST00000446558.1 |
RPL37A
|
ribosomal protein L37a |
| chr1_+_95975672 | 0.12 |
ENST00000440116.2
ENST00000456933.1 |
RP11-286B14.1
|
RP11-286B14.1 |
| chr11_+_92085262 | 0.12 |
ENST00000298047.6
ENST00000409404.2 ENST00000541502.1 |
FAT3
|
FAT atypical cadherin 3 |
| chr15_+_54305101 | 0.11 |
ENST00000260323.11
ENST00000545554.1 ENST00000537900.1 |
UNC13C
|
unc-13 homolog C (C. elegans) |
| chr13_-_103719196 | 0.11 |
ENST00000245312.3
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter), member 2 |
| chr19_-_42721819 | 0.11 |
ENST00000336034.4
ENST00000598200.1 ENST00000598727.1 ENST00000596251.1 |
DEDD2
|
death effector domain containing 2 |
| chr19_+_13122980 | 0.11 |
ENST00000590027.1
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr9_+_133971909 | 0.10 |
ENST00000247291.3
ENST00000372302.1 ENST00000372300.1 ENST00000372298.1 |
AIF1L
|
allograft inflammatory factor 1-like |
| chr3_+_159557637 | 0.10 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr5_+_169010638 | 0.10 |
ENST00000265295.4
ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr20_+_44330651 | 0.10 |
ENST00000305479.2
|
WFDC13
|
WAP four-disulfide core domain 13 |
| chr8_+_40010989 | 0.10 |
ENST00000315792.3
|
C8orf4
|
chromosome 8 open reading frame 4 |
| chr11_-_85376121 | 0.10 |
ENST00000527447.1
|
CREBZF
|
CREB/ATF bZIP transcription factor |
| chr6_+_131894284 | 0.09 |
ENST00000368087.3
ENST00000356962.2 |
ARG1
|
arginase 1 |
| chr10_-_37891859 | 0.09 |
ENST00000544824.1
|
MTRNR2L7
|
MT-RNR2-like 7 |
| chrM_+_10758 | 0.09 |
ENST00000361381.2
|
MT-ND4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr12_+_109785708 | 0.09 |
ENST00000310903.5
|
MYO1H
|
myosin IH |
| chr20_+_56964169 | 0.09 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr12_+_78359999 | 0.09 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr20_-_44600810 | 0.08 |
ENST00000322927.2
ENST00000426788.1 |
ZNF335
|
zinc finger protein 335 |
| chr7_-_83824169 | 0.08 |
ENST00000265362.4
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr17_-_60883993 | 0.08 |
ENST00000583803.1
ENST00000456609.2 |
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr2_-_106013400 | 0.08 |
ENST00000409807.1
|
FHL2
|
four and a half LIM domains 2 |
| chr7_-_115670804 | 0.08 |
ENST00000320239.7
|
TFEC
|
transcription factor EC |
| chr14_-_60043524 | 0.08 |
ENST00000537690.2
ENST00000281581.4 |
CCDC175
|
coiled-coil domain containing 175 |
| chr12_+_15699286 | 0.08 |
ENST00000442921.2
ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr8_+_107460147 | 0.08 |
ENST00000442977.2
|
OXR1
|
oxidation resistance 1 |
| chr2_+_201170770 | 0.08 |
ENST00000409988.3
ENST00000409385.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr3_+_174577070 | 0.07 |
ENST00000454872.1
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase-like 2 |
| chr12_+_60058458 | 0.07 |
ENST00000548610.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr1_+_36789335 | 0.07 |
ENST00000373137.2
|
RP11-268J15.5
|
RP11-268J15.5 |
| chr10_+_50507232 | 0.07 |
ENST00000374144.3
|
C10orf71
|
chromosome 10 open reading frame 71 |
| chr12_+_58176525 | 0.07 |
ENST00000543727.1
ENST00000540550.1 ENST00000323833.8 ENST00000350762.5 ENST00000550559.1 ENST00000548851.1 ENST00000434359.1 ENST00000457189.1 |
TSFM
|
Ts translation elongation factor, mitochondrial |
| chr3_+_14716606 | 0.07 |
ENST00000253697.3
ENST00000435614.1 ENST00000412910.1 |
C3orf20
|
chromosome 3 open reading frame 20 |
| chr8_-_100905850 | 0.07 |
ENST00000520271.1
ENST00000522940.1 ENST00000523016.1 ENST00000517682.2 ENST00000297564.2 |
COX6C
|
cytochrome c oxidase subunit VIc |
| chr1_+_247670415 | 0.07 |
ENST00000366491.2
ENST00000366489.1 ENST00000526896.1 |
GCSAML
|
germinal center-associated, signaling and motility-like |
| chr18_+_28956740 | 0.07 |
ENST00000308128.4
ENST00000359747.4 |
DSG4
|
desmoglein 4 |
| chr9_+_22646189 | 0.07 |
ENST00000436786.1
|
RP11-399D6.2
|
RP11-399D6.2 |
| chr7_-_6007070 | 0.06 |
ENST00000337579.3
|
RSPH10B
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chr10_-_32667660 | 0.06 |
ENST00000375110.2
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila) |
| chr5_+_140207536 | 0.06 |
ENST00000529310.1
ENST00000527624.1 |
PCDHA6
|
protocadherin alpha 6 |
| chr20_-_33413416 | 0.06 |
ENST00000359003.2
|
NCOA6
|
nuclear receptor coactivator 6 |
| chrM_+_7586 | 0.06 |
ENST00000361739.1
|
MT-CO2
|
mitochondrially encoded cytochrome c oxidase II |
| chr5_+_137225158 | 0.06 |
ENST00000290431.5
|
PKD2L2
|
polycystic kidney disease 2-like 2 |
| chr12_-_10251539 | 0.06 |
ENST00000420265.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr6_+_166945369 | 0.06 |
ENST00000598601.1
|
Z98049.1
|
CDNA FLJ25492 fis, clone CBR01389; Uncharacterized protein |
| chr5_+_140535577 | 0.06 |
ENST00000539533.1
|
PCDHB17
|
Protocadherin-psi1; Uncharacterized protein |
| chr9_+_133971863 | 0.06 |
ENST00000372309.3
|
AIF1L
|
allograft inflammatory factor 1-like |
| chr2_+_228337079 | 0.06 |
ENST00000409315.1
ENST00000373671.3 ENST00000409171.1 |
AGFG1
|
ArfGAP with FG repeats 1 |
| chr12_-_100656134 | 0.05 |
ENST00000548313.1
|
DEPDC4
|
DEP domain containing 4 |
| chrX_-_30595959 | 0.05 |
ENST00000378962.3
|
CXorf21
|
chromosome X open reading frame 21 |
| chr2_-_27435390 | 0.05 |
ENST00000428518.1
|
SLC5A6
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr7_-_115670792 | 0.05 |
ENST00000265440.7
ENST00000393485.1 |
TFEC
|
transcription factor EC |
| chr12_+_122880045 | 0.05 |
ENST00000539034.1
ENST00000535976.1 |
RP11-450K4.1
|
RP11-450K4.1 |
| chr19_+_19626531 | 0.05 |
ENST00000507754.4
|
NDUFA13
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13 |
| chr7_+_6796986 | 0.05 |
ENST00000297186.3
|
RSPH10B2
|
radial spoke head 10 homolog B2 (Chlamydomonas) |
| chr7_-_6006768 | 0.05 |
ENST00000441023.2
|
RSPH10B
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chr2_+_70121075 | 0.05 |
ENST00000409116.1
|
SNRNP27
|
small nuclear ribonucleoprotein 27kDa (U4/U6.U5) |
| chr5_-_78281623 | 0.05 |
ENST00000521117.1
|
ARSB
|
arylsulfatase B |
| chr8_-_126963417 | 0.05 |
ENST00000500989.2
|
LINC00861
|
long intergenic non-protein coding RNA 861 |
| chr14_+_85996507 | 0.05 |
ENST00000554746.1
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr7_+_6797288 | 0.05 |
ENST00000433859.2
ENST00000359718.3 |
RSPH10B2
|
radial spoke head 10 homolog B2 (Chlamydomonas) |
| chr5_-_39462390 | 0.05 |
ENST00000511792.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr14_-_90798418 | 0.04 |
ENST00000354366.3
|
NRDE2
|
NRDE-2, necessary for RNA interference, domain containing |
| chr3_-_37216055 | 0.04 |
ENST00000336686.4
|
LRRFIP2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr17_-_41623716 | 0.04 |
ENST00000319349.5
|
ETV4
|
ets variant 4 |
| chr4_+_71019903 | 0.04 |
ENST00000344526.5
|
C4orf40
|
chromosome 4 open reading frame 40 |
| chr4_-_66536057 | 0.04 |
ENST00000273854.3
|
EPHA5
|
EPH receptor A5 |
| chr8_+_18248755 | 0.04 |
ENST00000286479.3
|
NAT2
|
N-acetyltransferase 2 (arylamine N-acetyltransferase) |
| chr11_-_66313699 | 0.04 |
ENST00000526986.1
ENST00000310442.3 |
ZDHHC24
|
zinc finger, DHHC-type containing 24 |
| chr10_-_128975273 | 0.04 |
ENST00000424811.2
|
FAM196A
|
family with sequence similarity 196, member A |
| chr1_+_82165350 | 0.04 |
ENST00000359929.3
|
LPHN2
|
latrophilin 2 |
| chrX_-_114253536 | 0.04 |
ENST00000371936.1
|
IL13RA2
|
interleukin 13 receptor, alpha 2 |
| chr11_-_18062872 | 0.03 |
ENST00000250018.2
|
TPH1
|
tryptophan hydroxylase 1 |
| chr3_+_107318157 | 0.03 |
ENST00000406780.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chr12_+_21525818 | 0.03 |
ENST00000240652.3
ENST00000542023.1 ENST00000537593.1 |
IAPP
|
islet amyloid polypeptide |
| chr8_-_114449112 | 0.03 |
ENST00000455883.2
ENST00000352409.3 ENST00000297405.5 |
CSMD3
|
CUB and Sushi multiple domains 3 |
| chr11_-_72504681 | 0.03 |
ENST00000538536.1
ENST00000543304.1 ENST00000540587.1 ENST00000334805.6 |
STARD10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr12_-_49393092 | 0.03 |
ENST00000421952.2
|
DDN
|
dendrin |
| chr5_+_137203465 | 0.03 |
ENST00000239926.4
|
MYOT
|
myotilin |
| chr4_-_66536196 | 0.03 |
ENST00000511294.1
|
EPHA5
|
EPH receptor A5 |
| chr4_-_177190364 | 0.03 |
ENST00000296525.3
|
ASB5
|
ankyrin repeat and SOCS box containing 5 |
| chr3_-_112564797 | 0.03 |
ENST00000398214.1
ENST00000448932.1 |
CD200R1L
|
CD200 receptor 1-like |
| chr9_+_109625378 | 0.03 |
ENST00000277225.5
ENST00000457913.1 ENST00000472574.1 |
ZNF462
|
zinc finger protein 462 |
| chr21_-_39705286 | 0.03 |
ENST00000414189.1
|
AP001422.3
|
AP001422.3 |
| chrX_+_70503433 | 0.03 |
ENST00000276079.8
ENST00000373856.3 ENST00000373841.1 ENST00000420903.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chr20_+_56964253 | 0.03 |
ENST00000395802.3
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr16_-_28222797 | 0.03 |
ENST00000569951.1
ENST00000565698.1 |
XPO6
|
exportin 6 |
| chr11_-_72504637 | 0.02 |
ENST00000536377.1
ENST00000359373.5 |
STARD10
ARAP1
|
StAR-related lipid transfer (START) domain containing 10 ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr4_+_106473768 | 0.02 |
ENST00000265154.2
ENST00000420470.2 |
ARHGEF38
|
Rho guanine nucleotide exchange factor (GEF) 38 |
| chr3_-_125094093 | 0.02 |
ENST00000484491.1
ENST00000492394.1 ENST00000471196.1 ENST00000468369.1 ENST00000544464.1 ENST00000485866.1 ENST00000360647.4 |
ZNF148
|
zinc finger protein 148 |
| chr7_-_105319536 | 0.02 |
ENST00000477775.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr13_+_113699029 | 0.02 |
ENST00000423251.1
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr3_+_130648842 | 0.02 |
ENST00000508297.1
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr20_+_12989596 | 0.02 |
ENST00000434210.1
ENST00000399002.2 |
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr14_+_85996471 | 0.02 |
ENST00000330753.4
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr10_+_90346519 | 0.02 |
ENST00000371939.3
|
LIPJ
|
lipase, family member J |
| chr12_-_71031185 | 0.02 |
ENST00000548122.1
ENST00000551525.1 ENST00000550358.1 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr10_+_45495898 | 0.02 |
ENST00000298299.3
|
ZNF22
|
zinc finger protein 22 |
| chr19_-_44388116 | 0.01 |
ENST00000587539.1
|
ZNF404
|
zinc finger protein 404 |
| chr11_+_33061543 | 0.01 |
ENST00000432887.1
ENST00000528898.1 ENST00000531632.2 |
TCP11L1
|
t-complex 11, testis-specific-like 1 |
| chr5_+_173315283 | 0.01 |
ENST00000265085.5
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr16_+_20499024 | 0.01 |
ENST00000593357.1
|
AC137056.1
|
Uncharacterized protein; cDNA FLJ34659 fis, clone KIDNE2018863 |
| chr20_+_12989822 | 0.01 |
ENST00000378194.4
|
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr3_-_112693865 | 0.01 |
ENST00000471858.1
ENST00000295863.4 ENST00000308611.3 |
CD200R1
|
CD200 receptor 1 |
| chr10_+_121410882 | 0.01 |
ENST00000369085.3
|
BAG3
|
BCL2-associated athanogene 3 |
| chr11_+_10772534 | 0.01 |
ENST00000361367.2
|
CTR9
|
CTR9, Paf1/RNA polymerase II complex component |
| chr10_+_118349920 | 0.01 |
ENST00000531984.1
|
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr1_+_218458625 | 0.01 |
ENST00000366932.3
|
RRP15
|
ribosomal RNA processing 15 homolog (S. cerevisiae) |
| chr16_+_81772633 | 0.00 |
ENST00000566191.1
ENST00000565272.1 ENST00000563954.1 ENST00000565054.1 |
RP11-960L18.1
PLCG2
|
RP11-960L18.1 phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| chr14_-_23446900 | 0.00 |
ENST00000556731.1
|
AJUBA
|
ajuba LIM protein |
| chr1_-_11024258 | 0.00 |
ENST00000418570.2
|
C1orf127
|
chromosome 1 open reading frame 127 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF333
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0061580 | colon epithelial cell migration(GO:0061580) |
| 0.1 | 0.3 | GO:0099545 | trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.1 | 0.3 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.1 | 0.4 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.3 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.1 | 0.2 | GO:0002669 | tolerance induction dependent upon immune response(GO:0002461) positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.0 | 0.2 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 0.2 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.2 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0043091 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.0 | 0.1 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.3 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.3 | GO:0032433 | filopodium tip(GO:0032433) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 0.5 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.1 | 0.3 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 0.2 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.2 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.2 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.0 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.0 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |