Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
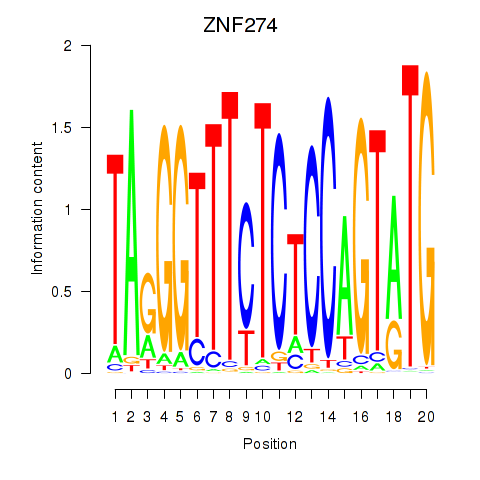
Results for ZNF274
Z-value: 1.10
Transcription factors associated with ZNF274
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF274
|
ENSG00000171606.13 | zinc finger protein 274 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF274 | hg19_v2_chr19_+_58694396_58694485 | 0.74 | 2.6e-01 | Click! |
Activity profile of ZNF274 motif
Sorted Z-values of ZNF274 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_40195101 | 1.20 |
ENST00000360675.3
ENST00000601802.1 |
LGALS14
|
lectin, galactoside-binding, soluble, 14 |
| chr2_-_96971259 | 0.79 |
ENST00000349783.5
|
SNRNP200
|
small nuclear ribonucleoprotein 200kDa (U5) |
| chr5_+_121465207 | 0.67 |
ENST00000296600.4
|
ZNF474
|
zinc finger protein 474 |
| chr15_+_89164520 | 0.67 |
ENST00000332810.3
|
AEN
|
apoptosis enhancing nuclease |
| chr12_-_49581152 | 0.62 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr2_+_9696028 | 0.54 |
ENST00000607241.1
|
RP11-214N9.1
|
RP11-214N9.1 |
| chr17_+_73028676 | 0.53 |
ENST00000581589.1
|
KCTD2
|
potassium channel tetramerization domain containing 2 |
| chr6_+_30687978 | 0.51 |
ENST00000327892.8
ENST00000435534.1 |
TUBB
|
tubulin, beta class I |
| chr19_-_56109119 | 0.50 |
ENST00000587678.1
|
FIZ1
|
FLT3-interacting zinc finger 1 |
| chr11_+_59705928 | 0.49 |
ENST00000398992.1
|
OOSP1
|
oocyte secreted protein 1 |
| chr3_+_37903432 | 0.48 |
ENST00000443503.2
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr17_-_41132410 | 0.47 |
ENST00000409446.3
ENST00000453594.1 ENST00000409399.1 ENST00000421990.2 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr16_+_29127282 | 0.46 |
ENST00000562902.1
|
RP11-426C22.5
|
RP11-426C22.5 |
| chr12_-_89919965 | 0.44 |
ENST00000548729.1
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough |
| chr9_+_125512019 | 0.44 |
ENST00000373684.1
ENST00000304720.2 |
OR1L6
|
olfactory receptor, family 1, subfamily L, member 6 |
| chr1_+_45205478 | 0.44 |
ENST00000452259.1
ENST00000372224.4 |
KIF2C
|
kinesin family member 2C |
| chr16_+_29819372 | 0.38 |
ENST00000568544.1
ENST00000569978.1 |
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr19_+_41949054 | 0.38 |
ENST00000378187.2
|
C19orf69
|
chromosome 19 open reading frame 69 |
| chrX_+_52920336 | 0.36 |
ENST00000452154.2
|
FAM156B
|
family with sequence similarity 156, member B |
| chr5_-_173043591 | 0.35 |
ENST00000285908.5
ENST00000480951.1 ENST00000311086.4 |
BOD1
|
biorientation of chromosomes in cell division 1 |
| chr1_+_45205498 | 0.35 |
ENST00000372218.4
|
KIF2C
|
kinesin family member 2C |
| chr15_+_89164560 | 0.35 |
ENST00000379231.3
ENST00000559528.1 |
AEN
|
apoptosis enhancing nuclease |
| chr6_+_31553978 | 0.33 |
ENST00000376096.1
ENST00000376099.1 ENST00000376110.3 |
LST1
|
leukocyte specific transcript 1 |
| chr16_+_31470179 | 0.31 |
ENST00000538189.1
ENST00000268314.4 |
ARMC5
|
armadillo repeat containing 5 |
| chr13_-_74993252 | 0.27 |
ENST00000325811.1
|
AL355390.1
|
Uncharacterized protein |
| chr5_-_74162605 | 0.23 |
ENST00000389156.4
ENST00000510496.1 ENST00000380515.3 |
FAM169A
|
family with sequence similarity 169, member A |
| chr16_+_31470143 | 0.22 |
ENST00000457010.2
ENST00000563544.1 |
ARMC5
|
armadillo repeat containing 5 |
| chr16_+_29818857 | 0.22 |
ENST00000567444.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chrX_+_119737806 | 0.19 |
ENST00000371317.5
|
MCTS1
|
malignant T cell amplified sequence 1 |
| chr7_-_73038822 | 0.18 |
ENST00000414749.2
ENST00000429400.2 ENST00000434326.1 |
MLXIPL
|
MLX interacting protein-like |
| chr17_+_41132564 | 0.18 |
ENST00000361677.1
ENST00000589705.1 |
RUNDC1
|
RUN domain containing 1 |
| chr22_+_39353527 | 0.17 |
ENST00000249116.2
|
APOBEC3A
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3A |
| chr20_+_60174827 | 0.16 |
ENST00000543233.1
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal) |
| chr1_-_114414316 | 0.15 |
ENST00000528414.1
ENST00000538253.1 ENST00000460620.1 ENST00000420377.2 ENST00000525799.1 ENST00000359785.5 |
PTPN22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr1_-_54355430 | 0.13 |
ENST00000371399.1
ENST00000072644.1 ENST00000412288.1 |
YIPF1
|
Yip1 domain family, member 1 |
| chr10_-_29811456 | 0.12 |
ENST00000535393.1
|
SVIL
|
supervillin |
| chr5_-_74162739 | 0.12 |
ENST00000513277.1
|
FAM169A
|
family with sequence similarity 169, member A |
| chr3_-_137893721 | 0.12 |
ENST00000505015.2
ENST00000260803.4 |
DBR1
|
debranching RNA lariats 1 |
| chr4_+_15704679 | 0.11 |
ENST00000382346.3
|
BST1
|
bone marrow stromal cell antigen 1 |
| chr1_+_42928945 | 0.11 |
ENST00000428554.2
|
CCDC30
|
coiled-coil domain containing 30 |
| chr12_-_89920030 | 0.09 |
ENST00000413530.1
ENST00000547474.1 |
GALNT4
POC1B-GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) POC1B-GALNT4 readthrough |
| chr11_+_16760161 | 0.09 |
ENST00000524439.1
ENST00000422258.2 ENST00000528634.1 ENST00000525684.1 |
C11orf58
|
chromosome 11 open reading frame 58 |
| chr7_+_30791743 | 0.09 |
ENST00000013222.5
ENST00000409539.1 |
INMT
|
indolethylamine N-methyltransferase |
| chr16_+_29819446 | 0.08 |
ENST00000568282.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr14_-_64846033 | 0.06 |
ENST00000556556.1
|
CTD-2555O16.1
|
CTD-2555O16.1 |
| chr13_+_103459704 | 0.05 |
ENST00000602836.1
|
BIVM-ERCC5
|
BIVM-ERCC5 readthrough |
| chr6_-_127780510 | 0.05 |
ENST00000487331.2
ENST00000483725.3 |
KIAA0408
|
KIAA0408 |
| chr4_+_15704573 | 0.04 |
ENST00000265016.4
|
BST1
|
bone marrow stromal cell antigen 1 |
| chr2_+_11864458 | 0.04 |
ENST00000396098.1
ENST00000396099.1 ENST00000425416.2 |
LPIN1
|
lipin 1 |
| chr3_+_50284321 | 0.03 |
ENST00000451956.1
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 |
| chr3_-_38992052 | 0.03 |
ENST00000302328.3
ENST00000450244.1 ENST00000444237.2 |
SCN11A
|
sodium channel, voltage-gated, type XI, alpha subunit |
| chr14_-_73997901 | 0.02 |
ENST00000557603.1
ENST00000556455.1 |
HEATR4
|
HEAT repeat containing 4 |
| chr10_+_5488564 | 0.02 |
ENST00000449083.1
ENST00000380359.3 |
NET1
|
neuroepithelial cell transforming 1 |
| chr19_+_7584088 | 0.01 |
ENST00000394341.2
|
ZNF358
|
zinc finger protein 358 |
| chr3_-_38991853 | 0.00 |
ENST00000456224.3
|
SCN11A
|
sodium channel, voltage-gated, type XI, alpha subunit |
| chr19_-_19774473 | 0.00 |
ENST00000357324.6
|
ATP13A1
|
ATPase type 13A1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF274
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.1 | 1.2 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.1 | 0.8 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.2 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.0 | 0.2 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.8 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.9 | GO:1990752 | microtubule end(GO:1990752) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.0 | 0.2 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.0 | 0.5 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.8 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |