Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
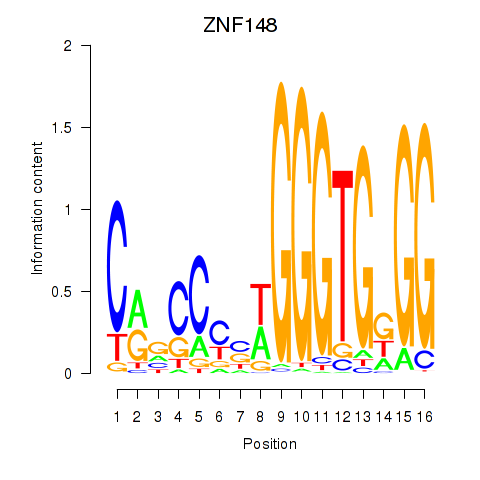
Results for ZNF148
Z-value: 0.41
Transcription factors associated with ZNF148
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF148
|
ENSG00000163848.14 | zinc finger protein 148 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF148 | hg19_v2_chr3_-_125094093_125094198 | -0.14 | 8.6e-01 | Click! |
Activity profile of ZNF148 motif
Sorted Z-values of ZNF148 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_172756506 | 0.30 |
ENST00000265087.4
|
STC2
|
stanniocalcin 2 |
| chr12_+_111843749 | 0.29 |
ENST00000341259.2
|
SH2B3
|
SH2B adaptor protein 3 |
| chr1_+_183155373 | 0.27 |
ENST00000493293.1
ENST00000264144.4 |
LAMC2
|
laminin, gamma 2 |
| chr19_+_10197463 | 0.26 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr11_-_33891362 | 0.25 |
ENST00000395833.3
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr19_+_18718214 | 0.23 |
ENST00000600490.1
|
TMEM59L
|
transmembrane protein 59-like |
| chr7_+_100770328 | 0.20 |
ENST00000223095.4
ENST00000445463.2 |
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr4_-_57976544 | 0.17 |
ENST00000295666.4
ENST00000537922.1 |
IGFBP7
|
insulin-like growth factor binding protein 7 |
| chr19_+_54371114 | 0.17 |
ENST00000448420.1
ENST00000439000.1 ENST00000391770.4 ENST00000391771.1 |
MYADM
|
myeloid-associated differentiation marker |
| chr20_-_47894569 | 0.17 |
ENST00000371744.1
ENST00000371752.1 ENST00000396105.1 |
ZNFX1
|
zinc finger, NFX1-type containing 1 |
| chr1_-_11714700 | 0.16 |
ENST00000354287.4
|
FBXO2
|
F-box protein 2 |
| chr13_-_44361025 | 0.15 |
ENST00000261488.6
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr1_+_150522222 | 0.15 |
ENST00000369039.5
|
ADAMTSL4
|
ADAMTS-like 4 |
| chr19_+_51815102 | 0.14 |
ENST00000270642.8
|
IGLON5
|
IgLON family member 5 |
| chr3_-_124560257 | 0.14 |
ENST00000496703.1
|
ITGB5
|
integrin, beta 5 |
| chr11_+_46402482 | 0.14 |
ENST00000441869.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr19_-_12267524 | 0.14 |
ENST00000455799.1
ENST00000355738.1 ENST00000439556.2 ENST00000542938.1 |
ZNF625
|
zinc finger protein 625 |
| chr10_-_47173994 | 0.14 |
ENST00000414655.2
ENST00000545298.1 ENST00000359178.4 ENST00000358140.4 ENST00000503031.1 |
ANXA8L1
LINC00842
|
annexin A8-like 1 long intergenic non-protein coding RNA 842 |
| chrX_+_48367338 | 0.14 |
ENST00000359882.4
ENST00000537758.1 ENST00000367574.4 ENST00000355961.4 ENST00000489940.1 ENST00000361988.3 |
PORCN
|
porcupine homolog (Drosophila) |
| chr5_-_141257954 | 0.14 |
ENST00000456271.1
ENST00000394536.3 ENST00000503492.1 ENST00000287008.3 |
PCDH1
|
protocadherin 1 |
| chr19_+_41725088 | 0.13 |
ENST00000301178.4
|
AXL
|
AXL receptor tyrosine kinase |
| chr11_+_46402297 | 0.13 |
ENST00000405308.2
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr12_-_54785054 | 0.13 |
ENST00000352268.6
ENST00000549962.1 |
ZNF385A
|
zinc finger protein 385A |
| chr12_+_38710555 | 0.13 |
ENST00000551464.1
|
ALG10B
|
ALG10B, alpha-1,2-glucosyltransferase |
| chr20_-_56285595 | 0.13 |
ENST00000395816.3
ENST00000347215.4 |
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr1_+_44401479 | 0.13 |
ENST00000438616.3
|
ARTN
|
artemin |
| chr4_+_75311019 | 0.12 |
ENST00000502307.1
|
AREG
|
amphiregulin |
| chr5_+_156887027 | 0.12 |
ENST00000435489.2
ENST00000311946.7 |
NIPAL4
|
NIPA-like domain containing 4 |
| chr1_+_20878932 | 0.12 |
ENST00000332947.4
|
FAM43B
|
family with sequence similarity 43, member B |
| chr12_-_54785074 | 0.12 |
ENST00000338010.5
ENST00000550774.1 |
ZNF385A
|
zinc finger protein 385A |
| chr19_+_41725140 | 0.12 |
ENST00000359092.3
|
AXL
|
AXL receptor tyrosine kinase |
| chr1_-_52456352 | 0.11 |
ENST00000371655.3
|
RAB3B
|
RAB3B, member RAS oncogene family |
| chr5_+_113769205 | 0.11 |
ENST00000503706.1
|
KCNN2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 |
| chr11_-_61735103 | 0.11 |
ENST00000529191.1
ENST00000529631.1 ENST00000530019.1 ENST00000529548.1 ENST00000273550.7 |
FTH1
|
ferritin, heavy polypeptide 1 |
| chr19_-_19049791 | 0.11 |
ENST00000594439.1
ENST00000221222.11 |
HOMER3
|
homer homolog 3 (Drosophila) |
| chr11_-_62313090 | 0.11 |
ENST00000528508.1
ENST00000533365.1 |
AHNAK
|
AHNAK nucleoprotein |
| chr6_+_30130969 | 0.11 |
ENST00000376694.4
|
TRIM15
|
tripartite motif containing 15 |
| chr2_-_47168850 | 0.11 |
ENST00000409207.1
|
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr1_+_150521876 | 0.11 |
ENST00000369041.5
ENST00000271643.4 ENST00000538795.1 |
ADAMTSL4
AL356356.1
|
ADAMTS-like 4 Protein LOC100996516 |
| chr4_+_75310851 | 0.10 |
ENST00000395748.3
ENST00000264487.2 |
AREG
|
amphiregulin |
| chr10_+_47746929 | 0.10 |
ENST00000340243.6
ENST00000374277.5 ENST00000449464.2 ENST00000538825.1 ENST00000335083.5 |
ANXA8L2
AL603965.1
|
annexin A8-like 2 Protein LOC100996760 |
| chr17_+_33914424 | 0.10 |
ENST00000590432.1
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr10_+_94608218 | 0.10 |
ENST00000371543.1
|
EXOC6
|
exocyst complex component 6 |
| chr19_+_10196781 | 0.10 |
ENST00000253110.11
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr6_-_31138439 | 0.10 |
ENST00000259915.8
|
POU5F1
|
POU class 5 homeobox 1 |
| chr15_-_59500973 | 0.10 |
ENST00000560749.1
|
MYO1E
|
myosin IE |
| chr11_-_61735029 | 0.10 |
ENST00000526640.1
|
FTH1
|
ferritin, heavy polypeptide 1 |
| chr10_+_48255253 | 0.10 |
ENST00000357718.4
ENST00000344416.5 ENST00000456111.2 ENST00000374258.3 |
ANXA8
AL591684.1
|
annexin A8 Protein LOC100996760 |
| chr17_+_33914276 | 0.10 |
ENST00000592545.1
ENST00000538556.1 ENST00000312678.8 ENST00000589344.1 |
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr2_-_220408260 | 0.10 |
ENST00000373891.2
|
CHPF
|
chondroitin polymerizing factor |
| chr16_+_330581 | 0.10 |
ENST00000219409.3
|
ARHGDIG
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr15_-_74494779 | 0.10 |
ENST00000571341.1
|
STRA6
|
stimulated by retinoic acid 6 |
| chr1_+_44445643 | 0.10 |
ENST00000309519.7
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr20_-_36793663 | 0.09 |
ENST00000536701.1
ENST00000536724.1 |
TGM2
|
transglutaminase 2 |
| chr12_+_6309517 | 0.09 |
ENST00000382519.4
ENST00000009180.4 |
CD9
|
CD9 molecule |
| chr11_+_1889880 | 0.09 |
ENST00000405957.2
|
LSP1
|
lymphocyte-specific protein 1 |
| chr6_-_31763276 | 0.09 |
ENST00000440048.1
|
VARS
|
valyl-tRNA synthetase |
| chr19_-_45579762 | 0.09 |
ENST00000303809.2
|
ZNF296
|
zinc finger protein 296 |
| chr15_-_74495188 | 0.09 |
ENST00000563965.1
ENST00000395105.4 |
STRA6
|
stimulated by retinoic acid 6 |
| chr20_-_44539538 | 0.09 |
ENST00000372420.1
|
PLTP
|
phospholipid transfer protein |
| chr11_-_61734599 | 0.09 |
ENST00000532601.1
|
FTH1
|
ferritin, heavy polypeptide 1 |
| chr19_-_663171 | 0.09 |
ENST00000606896.1
ENST00000589762.2 |
RNF126
|
ring finger protein 126 |
| chr8_-_135725205 | 0.09 |
ENST00000523399.1
ENST00000377838.3 |
ZFAT
|
zinc finger and AT hook domain containing |
| chr5_+_76114758 | 0.09 |
ENST00000514165.1
ENST00000296677.4 |
F2RL1
|
coagulation factor II (thrombin) receptor-like 1 |
| chr1_-_95007193 | 0.09 |
ENST00000370207.4
ENST00000334047.7 |
F3
|
coagulation factor III (thromboplastin, tissue factor) |
| chr3_+_41241596 | 0.09 |
ENST00000450969.1
|
CTNNB1
|
catenin (cadherin-associated protein), beta 1, 88kDa |
| chr17_+_33914460 | 0.09 |
ENST00000537622.2
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr3_+_49977894 | 0.09 |
ENST00000433811.1
|
RBM6
|
RNA binding motif protein 6 |
| chr9_+_19049372 | 0.09 |
ENST00000380527.1
|
RRAGA
|
Ras-related GTP binding A |
| chr2_+_220144168 | 0.09 |
ENST00000392087.2
ENST00000442681.1 ENST00000439026.1 |
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr4_+_75480629 | 0.09 |
ENST00000380846.3
|
AREGB
|
amphiregulin B |
| chr19_-_11616645 | 0.09 |
ENST00000293771.5
|
ZNF653
|
zinc finger protein 653 |
| chr11_+_66742742 | 0.09 |
ENST00000308963.4
|
C11orf86
|
chromosome 11 open reading frame 86 |
| chr5_-_176037105 | 0.08 |
ENST00000303991.4
|
GPRIN1
|
G protein regulated inducer of neurite outgrowth 1 |
| chr15_+_66994663 | 0.08 |
ENST00000457357.2
|
SMAD6
|
SMAD family member 6 |
| chr2_-_47572207 | 0.08 |
ENST00000441997.1
|
AC073283.4
|
AC073283.4 |
| chr11_+_67806467 | 0.08 |
ENST00000265686.3
ENST00000524598.1 ENST00000529657.1 |
TCIRG1
|
T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 subunit A3 |
| chr1_-_201346761 | 0.08 |
ENST00000455702.1
ENST00000422165.1 ENST00000367318.5 ENST00000367320.2 ENST00000438742.1 ENST00000412633.1 ENST00000458432.2 ENST00000421663.2 ENST00000367322.1 ENST00000509001.1 |
TNNT2
|
troponin T type 2 (cardiac) |
| chr17_+_7348658 | 0.08 |
ENST00000570557.1
ENST00000536404.2 ENST00000576360.1 |
CHRNB1
|
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr3_+_183415558 | 0.08 |
ENST00000305135.5
|
YEATS2
|
YEATS domain containing 2 |
| chr16_+_57139933 | 0.08 |
ENST00000566259.1
|
CPNE2
|
copine II |
| chr22_-_20256054 | 0.08 |
ENST00000043402.7
|
RTN4R
|
reticulon 4 receptor |
| chr6_-_29648887 | 0.08 |
ENST00000376883.1
|
ZFP57
|
ZFP57 zinc finger protein |
| chr16_+_58035277 | 0.08 |
ENST00000219281.3
ENST00000539737.2 ENST00000423271.3 ENST00000561568.1 ENST00000563149.1 |
USB1
|
U6 snRNA biogenesis 1 |
| chr17_+_46125685 | 0.08 |
ENST00000579889.1
|
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr17_+_7348374 | 0.08 |
ENST00000306071.2
ENST00000572857.1 |
CHRNB1
|
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr6_+_144471643 | 0.08 |
ENST00000367568.4
|
STX11
|
syntaxin 11 |
| chr19_+_54369608 | 0.08 |
ENST00000336967.3
|
MYADM
|
myeloid-associated differentiation marker |
| chr10_+_75532028 | 0.08 |
ENST00000372841.3
ENST00000394790.1 |
FUT11
|
fucosyltransferase 11 (alpha (1,3) fucosyltransferase) |
| chr8_-_123793048 | 0.08 |
ENST00000607710.1
|
RP11-44N11.2
|
RP11-44N11.2 |
| chr10_+_90672113 | 0.08 |
ENST00000371922.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr16_-_776431 | 0.08 |
ENST00000293889.6
|
CCDC78
|
coiled-coil domain containing 78 |
| chr17_-_47308128 | 0.08 |
ENST00000413580.1
ENST00000511066.1 |
PHOSPHO1
|
phosphatase, orphan 1 |
| chr3_-_49131614 | 0.08 |
ENST00000450685.1
|
QRICH1
|
glutamine-rich 1 |
| chr21_+_45161301 | 0.08 |
ENST00000467908.1
|
PDXK
|
pyridoxal (pyridoxine, vitamin B6) kinase |
| chrX_+_73641286 | 0.07 |
ENST00000587091.1
|
SLC16A2
|
solute carrier family 16, member 2 (thyroid hormone transporter) |
| chr15_+_66994561 | 0.07 |
ENST00000288840.5
|
SMAD6
|
SMAD family member 6 |
| chr11_-_118661588 | 0.07 |
ENST00000534980.1
ENST00000526070.2 |
DDX6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr20_-_4804244 | 0.07 |
ENST00000379400.3
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2 |
| chr20_-_36793774 | 0.07 |
ENST00000361475.2
|
TGM2
|
transglutaminase 2 |
| chr14_-_82089405 | 0.07 |
ENST00000554211.1
|
RP11-799P8.1
|
RP11-799P8.1 |
| chr10_-_73533255 | 0.07 |
ENST00000394957.3
|
C10orf54
|
chromosome 10 open reading frame 54 |
| chr12_+_54694979 | 0.07 |
ENST00000552848.1
|
COPZ1
|
coatomer protein complex, subunit zeta 1 |
| chr18_+_21464737 | 0.07 |
ENST00000586751.1
|
LAMA3
|
laminin, alpha 3 |
| chr17_-_42992856 | 0.07 |
ENST00000588316.1
ENST00000435360.2 ENST00000586793.1 ENST00000588735.1 ENST00000588037.1 ENST00000592320.1 ENST00000253408.5 |
GFAP
|
glial fibrillary acidic protein |
| chr17_-_27278445 | 0.07 |
ENST00000268756.3
ENST00000584685.1 |
PHF12
|
PHD finger protein 12 |
| chr6_-_31651817 | 0.07 |
ENST00000375863.3
ENST00000375860.2 |
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C |
| chr14_-_69444957 | 0.07 |
ENST00000556571.1
ENST00000553659.1 ENST00000555616.1 |
ACTN1
|
actinin, alpha 1 |
| chr22_-_30662828 | 0.07 |
ENST00000403463.1
ENST00000215781.2 |
OSM
|
oncostatin M |
| chr11_-_45687128 | 0.07 |
ENST00000308064.2
|
CHST1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 |
| chr3_-_88108212 | 0.07 |
ENST00000482016.1
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr11_+_3666335 | 0.07 |
ENST00000250693.1
|
ART1
|
ADP-ribosyltransferase 1 |
| chr2_+_85132749 | 0.07 |
ENST00000233143.4
|
TMSB10
|
thymosin beta 10 |
| chr19_+_44037334 | 0.07 |
ENST00000314228.5
|
ZNF575
|
zinc finger protein 575 |
| chr17_+_36508111 | 0.07 |
ENST00000331159.5
ENST00000577233.1 |
SOCS7
|
suppressor of cytokine signaling 7 |
| chrX_-_152989798 | 0.07 |
ENST00000441714.1
ENST00000442093.1 ENST00000429550.1 ENST00000345046.6 |
BCAP31
|
B-cell receptor-associated protein 31 |
| chr19_+_7599597 | 0.07 |
ENST00000414982.3
ENST00000450331.3 |
PNPLA6
|
patatin-like phospholipase domain containing 6 |
| chr1_-_47082495 | 0.06 |
ENST00000545730.1
ENST00000531769.1 ENST00000319928.3 |
MKNK1
MOB3C
|
MAP kinase interacting serine/threonine kinase 1 MOB kinase activator 3C |
| chr22_+_35653445 | 0.06 |
ENST00000420166.1
ENST00000444518.2 ENST00000455359.1 ENST00000216106.5 |
HMGXB4
|
HMG box domain containing 4 |
| chr2_+_220144052 | 0.06 |
ENST00000425450.1
ENST00000392086.4 ENST00000421532.1 |
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr6_-_13814663 | 0.06 |
ENST00000359495.2
ENST00000379170.4 |
MCUR1
|
mitochondrial calcium uniporter regulator 1 |
| chr8_+_104383759 | 0.06 |
ENST00000415886.2
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr2_+_128848881 | 0.06 |
ENST00000259253.6
|
UGGT1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr10_+_99258625 | 0.06 |
ENST00000370664.3
|
UBTD1
|
ubiquitin domain containing 1 |
| chr1_-_89738528 | 0.06 |
ENST00000343435.5
|
GBP5
|
guanylate binding protein 5 |
| chr8_+_145065705 | 0.06 |
ENST00000533044.1
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr2_+_16080659 | 0.06 |
ENST00000281043.3
|
MYCN
|
v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog |
| chr10_-_30348439 | 0.06 |
ENST00000375377.1
|
KIAA1462
|
KIAA1462 |
| chr17_+_53828333 | 0.06 |
ENST00000268896.5
|
PCTP
|
phosphatidylcholine transfer protein |
| chr18_+_1099004 | 0.06 |
ENST00000581556.1
|
RP11-78F17.1
|
RP11-78F17.1 |
| chr2_-_47168906 | 0.06 |
ENST00000444761.2
ENST00000409147.1 |
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr15_-_74043816 | 0.06 |
ENST00000379822.4
|
C15orf59
|
chromosome 15 open reading frame 59 |
| chr19_+_42746927 | 0.06 |
ENST00000378108.1
|
AC006486.1
|
AC006486.1 |
| chr1_-_1208851 | 0.06 |
ENST00000488418.1
|
UBE2J2
|
ubiquitin-conjugating enzyme E2, J2 |
| chr21_+_44394742 | 0.06 |
ENST00000432907.2
|
PKNOX1
|
PBX/knotted 1 homeobox 1 |
| chr2_+_220306745 | 0.06 |
ENST00000431523.1
ENST00000396698.1 ENST00000396695.2 |
SPEG
|
SPEG complex locus |
| chr12_-_117799446 | 0.06 |
ENST00000317775.6
ENST00000344089.3 |
NOS1
|
nitric oxide synthase 1 (neuronal) |
| chrX_+_107069063 | 0.06 |
ENST00000262843.6
|
MID2
|
midline 2 |
| chr19_+_54369434 | 0.06 |
ENST00000421337.1
|
MYADM
|
myeloid-associated differentiation marker |
| chr3_+_57994127 | 0.05 |
ENST00000490882.1
ENST00000295956.4 ENST00000358537.3 ENST00000429972.2 ENST00000348383.5 ENST00000357272.4 |
FLNB
|
filamin B, beta |
| chr19_+_10196981 | 0.05 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr2_+_153191706 | 0.05 |
ENST00000288670.9
|
FMNL2
|
formin-like 2 |
| chr17_-_56065540 | 0.05 |
ENST00000583932.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr17_-_46507567 | 0.05 |
ENST00000584924.1
|
SKAP1
|
src kinase associated phosphoprotein 1 |
| chr9_-_37904084 | 0.05 |
ENST00000377716.2
ENST00000242275.6 |
SLC25A51
|
solute carrier family 25, member 51 |
| chr4_-_6675550 | 0.05 |
ENST00000513179.1
ENST00000515205.1 |
RP11-539L10.3
|
RP11-539L10.3 |
| chr1_-_155224699 | 0.05 |
ENST00000491082.1
|
FAM189B
|
family with sequence similarity 189, member B |
| chr9_+_35538616 | 0.05 |
ENST00000455600.1
|
RUSC2
|
RUN and SH3 domain containing 2 |
| chr14_+_24837226 | 0.05 |
ENST00000554050.1
ENST00000554903.1 ENST00000554779.1 ENST00000250373.4 ENST00000553708.1 |
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 |
| chr9_+_95858485 | 0.05 |
ENST00000375464.2
|
C9orf89
|
chromosome 9 open reading frame 89 |
| chr17_+_38337491 | 0.05 |
ENST00000538981.1
|
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr2_+_201171372 | 0.05 |
ENST00000409140.3
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr7_+_100273736 | 0.05 |
ENST00000412215.1
ENST00000393924.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr2_-_136873735 | 0.05 |
ENST00000409817.1
|
CXCR4
|
chemokine (C-X-C motif) receptor 4 |
| chr7_-_80548493 | 0.05 |
ENST00000536800.1
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr2_+_128848740 | 0.05 |
ENST00000375990.3
|
UGGT1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr1_+_204485571 | 0.05 |
ENST00000454264.2
ENST00000367183.3 |
MDM4
|
Mdm4 p53 binding protein homolog (mouse) |
| chr19_+_797392 | 0.05 |
ENST00000350092.4
ENST00000349038.4 ENST00000586481.1 ENST00000585535.1 |
PTBP1
|
polypyrimidine tract binding protein 1 |
| chr1_+_145576007 | 0.05 |
ENST00000369298.1
|
PIAS3
|
protein inhibitor of activated STAT, 3 |
| chr12_+_132312931 | 0.05 |
ENST00000360564.1
ENST00000545671.1 ENST00000545790.1 |
MMP17
|
matrix metallopeptidase 17 (membrane-inserted) |
| chr17_+_2699697 | 0.05 |
ENST00000254695.8
ENST00000366401.4 ENST00000542807.1 |
RAP1GAP2
|
RAP1 GTPase activating protein 2 |
| chr17_+_41177220 | 0.05 |
ENST00000587250.2
ENST00000544533.1 |
RND2
|
Rho family GTPase 2 |
| chr11_-_118661828 | 0.05 |
ENST00000264018.4
|
DDX6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr2_-_61697862 | 0.05 |
ENST00000398571.2
|
USP34
|
ubiquitin specific peptidase 34 |
| chr11_+_34938119 | 0.05 |
ENST00000227868.4
ENST00000430469.2 ENST00000533262.1 |
PDHX
|
pyruvate dehydrogenase complex, component X |
| chr16_+_29984962 | 0.05 |
ENST00000308893.4
|
TAOK2
|
TAO kinase 2 |
| chr16_+_2732476 | 0.05 |
ENST00000301738.4
ENST00000564195.1 |
KCTD5
|
potassium channel tetramerization domain containing 5 |
| chr1_-_201476220 | 0.05 |
ENST00000526723.1
ENST00000524951.1 |
CSRP1
|
cysteine and glycine-rich protein 1 |
| chr1_+_145575980 | 0.05 |
ENST00000393045.2
|
PIAS3
|
protein inhibitor of activated STAT, 3 |
| chr2_+_220143989 | 0.05 |
ENST00000336576.5
|
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr2_-_220408430 | 0.05 |
ENST00000243776.6
|
CHPF
|
chondroitin polymerizing factor |
| chr2_-_165477971 | 0.05 |
ENST00000446413.2
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr9_+_130565487 | 0.05 |
ENST00000373225.3
ENST00000431857.1 |
FPGS
|
folylpolyglutamate synthase |
| chr1_-_113161730 | 0.05 |
ENST00000544629.1
ENST00000543570.1 ENST00000360743.4 ENST00000490067.1 ENST00000343210.7 ENST00000369666.1 |
ST7L
|
suppression of tumorigenicity 7 like |
| chr20_+_55205825 | 0.05 |
ENST00000544508.1
|
TFAP2C
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr8_-_91618285 | 0.05 |
ENST00000517505.1
|
LINC01030
|
long intergenic non-protein coding RNA 1030 |
| chr19_-_10047219 | 0.05 |
ENST00000264833.4
|
OLFM2
|
olfactomedin 2 |
| chr1_+_40506255 | 0.04 |
ENST00000421589.1
|
CAP1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr8_-_25315905 | 0.04 |
ENST00000221200.4
|
KCTD9
|
potassium channel tetramerization domain containing 9 |
| chr17_-_34890759 | 0.04 |
ENST00000431794.3
|
MYO19
|
myosin XIX |
| chr1_-_153917700 | 0.04 |
ENST00000368646.2
|
DENND4B
|
DENN/MADD domain containing 4B |
| chr6_-_31633624 | 0.04 |
ENST00000375895.2
ENST00000375900.4 |
GPANK1
|
G patch domain and ankyrin repeats 1 |
| chr6_-_38607673 | 0.04 |
ENST00000481247.1
|
BTBD9
|
BTB (POZ) domain containing 9 |
| chr6_+_15401075 | 0.04 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr19_+_12944722 | 0.04 |
ENST00000591495.1
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chr16_+_89894875 | 0.04 |
ENST00000393062.2
|
SPIRE2
|
spire-type actin nucleation factor 2 |
| chr22_+_30279144 | 0.04 |
ENST00000401950.2
ENST00000333027.3 ENST00000445401.1 ENST00000323630.5 ENST00000351488.3 |
MTMR3
|
myotubularin related protein 3 |
| chr18_-_33077868 | 0.04 |
ENST00000590757.1
ENST00000592173.1 ENST00000441607.2 ENST00000587450.1 ENST00000589258.1 |
INO80C
RP11-322E11.6
|
INO80 complex subunit C Uncharacterized protein |
| chr1_+_40506392 | 0.04 |
ENST00000414893.1
ENST00000414281.1 ENST00000420216.1 ENST00000372792.2 ENST00000372798.1 ENST00000340450.3 ENST00000372805.3 ENST00000435719.1 ENST00000427843.1 ENST00000417287.1 ENST00000424977.1 ENST00000446031.1 |
CAP1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr16_+_2213530 | 0.04 |
ENST00000567645.1
|
TRAF7
|
TNF receptor-associated factor 7, E3 ubiquitin protein ligase |
| chr2_+_173292059 | 0.04 |
ENST00000412899.1
ENST00000409532.1 |
ITGA6
|
integrin, alpha 6 |
| chr17_-_40540484 | 0.04 |
ENST00000588969.1
|
STAT3
|
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr19_+_7600584 | 0.04 |
ENST00000600737.1
|
PNPLA6
|
patatin-like phospholipase domain containing 6 |
| chr2_+_239335636 | 0.04 |
ENST00000409297.1
|
ASB1
|
ankyrin repeat and SOCS box containing 1 |
| chr12_+_56522001 | 0.04 |
ENST00000267113.4
ENST00000541590.1 |
ESYT1
|
extended synaptotagmin-like protein 1 |
| chr9_-_4679419 | 0.04 |
ENST00000609131.1
ENST00000607997.1 |
RP11-6J24.6
|
RP11-6J24.6 |
| chr1_+_202995611 | 0.04 |
ENST00000367240.2
|
PPFIA4
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 |
| chr1_+_116915270 | 0.04 |
ENST00000418797.1
|
ATP1A1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
| chr1_-_159893507 | 0.04 |
ENST00000368096.1
|
TAGLN2
|
transgelin 2 |
| chr18_-_33077556 | 0.04 |
ENST00000589273.1
ENST00000586489.1 |
INO80C
|
INO80 complex subunit C |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF148
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.1 | 0.3 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.3 | GO:2000669 | negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 0.0 | 0.2 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.0 | 0.2 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.3 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.3 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.1 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.0 | 0.3 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.0 | 0.2 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.2 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.0 | 0.1 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.1 | GO:0071629 | cytoplasm-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071629) |
| 0.0 | 0.1 | GO:0043311 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.2 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 | 0.1 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.1 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.0 | 0.1 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 0.0 | 0.2 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:1904793 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.0 | 0.1 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.0 | 0.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.1 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.0 | 0.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.1 | GO:0072616 | interleukin-18 secretion(GO:0072616) |
| 0.0 | 0.3 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.3 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.0 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.0 | 0.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.1 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.3 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 | 0.3 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.0 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.1 | 0.3 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.0 | 0.1 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.1 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.3 | GO:0033643 | host cell part(GO:0033643) |
| 0.0 | 0.1 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0070369 | Scrib-APC-beta-catenin complex(GO:0034750) beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.3 | GO:0036020 | endolysosome membrane(GO:0036020) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070698 | transforming growth factor beta receptor, inhibitory cytoplasmic mediator activity(GO:0030617) type I activin receptor binding(GO:0070698) |
| 0.0 | 0.1 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.1 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 0.1 | GO:0008841 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) dihydrofolate synthase activity(GO:0008841) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.0 | 0.3 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.0 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |