Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
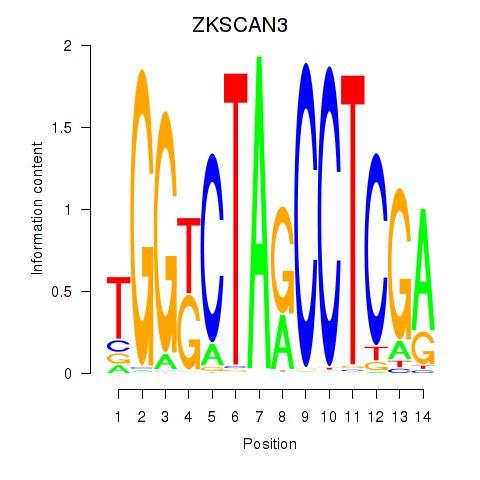
Results for ZKSCAN3
Z-value: 1.43
Transcription factors associated with ZKSCAN3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZKSCAN3
|
ENSG00000189298.9 | zinc finger with KRAB and SCAN domains 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZKSCAN3 | hg19_v2_chr6_+_28317685_28317761 | -0.39 | 6.1e-01 | Click! |
Activity profile of ZKSCAN3 motif
Sorted Z-values of ZKSCAN3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_39786962 | 11.72 |
ENST00000333625.2
|
IFNL1
|
interferon, lambda 1 |
| chr19_-_39735646 | 2.10 |
ENST00000413851.2
|
IFNL3
|
interferon, lambda 3 |
| chr19_+_39759154 | 1.90 |
ENST00000331982.5
|
IFNL2
|
interferon, lambda 2 |
| chr1_-_935361 | 1.15 |
ENST00000484667.2
|
HES4
|
hes family bHLH transcription factor 4 |
| chr12_-_3982511 | 0.94 |
ENST00000427057.2
ENST00000228820.4 |
PARP11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr12_-_3982548 | 0.68 |
ENST00000397096.2
ENST00000447133.3 ENST00000450737.2 |
PARP11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr7_+_1084206 | 0.59 |
ENST00000444847.1
|
GPR146
|
G protein-coupled receptor 146 |
| chr7_-_155089251 | 0.56 |
ENST00000609974.1
|
AC144652.1
|
AC144652.1 |
| chr19_-_4182530 | 0.56 |
ENST00000601571.1
ENST00000601488.1 ENST00000305232.6 ENST00000381935.3 ENST00000337491.2 |
SIRT6
|
sirtuin 6 |
| chr9_+_127023704 | 0.55 |
ENST00000373596.1
ENST00000425237.1 |
NEK6
|
NIMA-related kinase 6 |
| chr17_+_32597232 | 0.54 |
ENST00000378569.2
ENST00000200307.4 ENST00000394627.1 ENST00000394630.3 |
CCL7
|
chemokine (C-C motif) ligand 7 |
| chr19_+_54372639 | 0.52 |
ENST00000391769.2
|
MYADM
|
myeloid-associated differentiation marker |
| chr19_-_4182497 | 0.48 |
ENST00000597896.1
|
SIRT6
|
sirtuin 6 |
| chr12_+_113796347 | 0.47 |
ENST00000545182.2
ENST00000280800.3 |
PLBD2
|
phospholipase B domain containing 2 |
| chr2_+_191745535 | 0.46 |
ENST00000320717.3
|
GLS
|
glutaminase |
| chr5_-_180235755 | 0.43 |
ENST00000502678.1
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr18_-_74728998 | 0.41 |
ENST00000359645.3
ENST00000397875.3 ENST00000397869.3 ENST00000578193.1 ENST00000578873.1 ENST00000397866.4 ENST00000528160.1 ENST00000527041.1 ENST00000526111.1 ENST00000397865.5 ENST00000382582.3 |
MBP
|
myelin basic protein |
| chr1_+_114472992 | 0.35 |
ENST00000514621.1
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr15_+_74610894 | 0.35 |
ENST00000558821.1
ENST00000268082.4 |
CCDC33
|
coiled-coil domain containing 33 |
| chr11_+_66610883 | 0.35 |
ENST00000309657.3
ENST00000524506.1 |
RCE1
|
Ras converting CAAX endopeptidase 1 |
| chr17_+_36908984 | 0.34 |
ENST00000225426.4
ENST00000579088.1 |
PSMB3
|
proteasome (prosome, macropain) subunit, beta type, 3 |
| chr1_+_114473350 | 0.33 |
ENST00000503968.1
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr4_-_169239921 | 0.31 |
ENST00000514995.1
ENST00000393743.3 |
DDX60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr16_-_1429010 | 0.31 |
ENST00000513783.1
|
UNKL
|
unkempt family zinc finger-like |
| chr1_+_3598871 | 0.26 |
ENST00000603362.1
ENST00000604479.1 |
TP73
|
tumor protein p73 |
| chrX_-_153775426 | 0.26 |
ENST00000393562.2
|
G6PD
|
glucose-6-phosphate dehydrogenase |
| chr19_+_32836499 | 0.25 |
ENST00000311921.4
ENST00000544431.1 ENST00000355898.5 |
ZNF507
|
zinc finger protein 507 |
| chr19_-_42348692 | 0.24 |
ENST00000330743.3
ENST00000601246.1 |
LYPD4
|
LY6/PLAUR domain containing 4 |
| chr2_-_61244308 | 0.23 |
ENST00000407787.1
ENST00000398658.2 |
PUS10
|
pseudouridylate synthase 10 |
| chr1_-_19811996 | 0.20 |
ENST00000264203.3
ENST00000401084.2 |
CAPZB
|
capping protein (actin filament) muscle Z-line, beta |
| chr1_-_86622421 | 0.20 |
ENST00000370571.2
|
COL24A1
|
collagen, type XXIV, alpha 1 |
| chr5_-_95297534 | 0.19 |
ENST00000513343.1
ENST00000431061.2 |
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr6_+_116850174 | 0.17 |
ENST00000416171.2
ENST00000368597.2 ENST00000452373.1 ENST00000405399.1 |
FAM26D
|
family with sequence similarity 26, member D |
| chr21_+_39668831 | 0.13 |
ENST00000419868.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr5_+_140079919 | 0.12 |
ENST00000274712.3
|
ZMAT2
|
zinc finger, matrin-type 2 |
| chr21_+_39668478 | 0.12 |
ENST00000398927.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chrX_+_23352133 | 0.11 |
ENST00000379361.4
|
PTCHD1
|
patched domain containing 1 |
| chr12_+_109535923 | 0.11 |
ENST00000336865.2
|
UNG
|
uracil-DNA glycosylase |
| chr19_+_52693259 | 0.11 |
ENST00000322088.6
ENST00000454220.2 ENST00000444322.2 ENST00000477989.1 |
PPP2R1A
|
protein phosphatase 2, regulatory subunit A, alpha |
| chr2_-_175351744 | 0.11 |
ENST00000295500.4
ENST00000392552.2 ENST00000392551.2 |
GPR155
|
G protein-coupled receptor 155 |
| chr16_-_745946 | 0.10 |
ENST00000562563.1
|
FBXL16
|
F-box and leucine-rich repeat protein 16 |
| chr11_-_67272794 | 0.09 |
ENST00000436757.2
ENST00000356404.3 |
PITPNM1
|
phosphatidylinositol transfer protein, membrane-associated 1 |
| chr16_-_832054 | 0.08 |
ENST00000543963.1
|
MSLNL
|
mesothelin-like |
| chrX_+_153775821 | 0.07 |
ENST00000263518.6
ENST00000470142.1 ENST00000393549.2 ENST00000455588.2 ENST00000369602.3 |
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr16_-_832926 | 0.07 |
ENST00000293892.3
|
MSLNL
|
mesothelin-like |
| chr17_-_60142609 | 0.07 |
ENST00000397786.2
|
MED13
|
mediator complex subunit 13 |
| chr6_-_31508304 | 0.06 |
ENST00000376177.2
|
DDX39B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr16_-_4039001 | 0.05 |
ENST00000576936.1
|
ADCY9
|
adenylate cyclase 9 |
| chr22_+_50312379 | 0.04 |
ENST00000407217.3
ENST00000403427.3 |
CRELD2
|
cysteine-rich with EGF-like domains 2 |
| chr14_-_106926724 | 0.04 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr5_-_65017921 | 0.04 |
ENST00000381007.4
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr11_-_61062762 | 0.04 |
ENST00000335613.5
|
VWCE
|
von Willebrand factor C and EGF domains |
| chr1_-_28415204 | 0.03 |
ENST00000373871.3
|
EYA3
|
eyes absent homolog 3 (Drosophila) |
| chr19_-_49258606 | 0.02 |
ENST00000310160.3
|
FUT1
|
fucosyltransferase 1 (galactoside 2-alpha-L-fucosyltransferase, H blood group) |
| chr1_-_19811132 | 0.02 |
ENST00000433834.1
|
CAPZB
|
capping protein (actin filament) muscle Z-line, beta |
| chr6_+_32936353 | 0.01 |
ENST00000374825.4
|
BRD2
|
bromodomain containing 2 |
| chr22_+_50312274 | 0.00 |
ENST00000404488.3
|
CRELD2
|
cysteine-rich with EGF-like domains 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZKSCAN3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 11.7 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.3 | 1.0 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) histone H3-K9 deacetylation(GO:1990619) |
| 0.1 | 0.3 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.1 | 0.4 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.1 | 0.5 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 1.9 | GO:0002385 | mucosal immune response(GO:0002385) |
| 0.1 | 0.5 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.3 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.7 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.4 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.2 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.2 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 2.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.6 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.0 | 0.3 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.7 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.3 | 1.0 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.4 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.3 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 1.6 | GO:0005643 | nuclear pore(GO:0005643) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.1 | 0.5 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 15.7 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.4 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 1.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.6 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |