Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
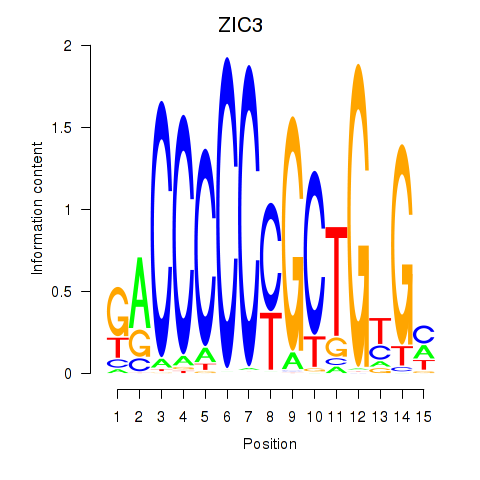
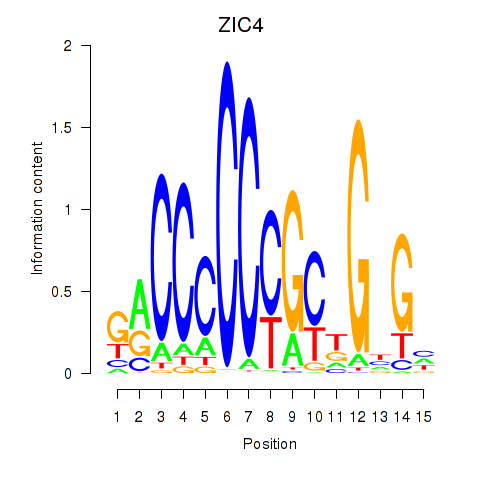
Results for ZIC3_ZIC4
Z-value: 0.76
Transcription factors associated with ZIC3_ZIC4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZIC3
|
ENSG00000156925.7 | Zic family member 3 |
|
ZIC4
|
ENSG00000174963.13 | Zic family member 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZIC3 | hg19_v2_chrX_+_136648643_136648711 | 0.96 | 4.3e-02 | Click! |
Activity profile of ZIC3_ZIC4 motif
Sorted Z-values of ZIC3_ZIC4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_302918 | 0.95 |
ENST00000599994.1
|
AC187652.1
|
Protein LOC100996433 |
| chr8_-_145060593 | 0.71 |
ENST00000313059.5
ENST00000524918.1 ENST00000313028.7 ENST00000525773.1 |
PARP10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr20_+_34203794 | 0.64 |
ENST00000374273.3
|
SPAG4
|
sperm associated antigen 4 |
| chr17_-_43045439 | 0.62 |
ENST00000253407.3
|
C1QL1
|
complement component 1, q subcomponent-like 1 |
| chr20_+_61340179 | 0.48 |
ENST00000370501.3
|
NTSR1
|
neurotensin receptor 1 (high affinity) |
| chr12_-_54779511 | 0.47 |
ENST00000551109.1
ENST00000546970.1 |
ZNF385A
|
zinc finger protein 385A |
| chr2_+_8822113 | 0.41 |
ENST00000396290.1
ENST00000331129.3 |
ID2
|
inhibitor of DNA binding 2, dominant negative helix-loop-helix protein |
| chr19_-_31840130 | 0.40 |
ENST00000558569.1
|
TSHZ3
|
teashirt zinc finger homeobox 3 |
| chr10_-_79397547 | 0.38 |
ENST00000481070.1
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr3_+_38179969 | 0.37 |
ENST00000396334.3
ENST00000417037.2 ENST00000424893.1 ENST00000495303.1 ENST00000443433.2 ENST00000421516.1 |
MYD88
|
myeloid differentiation primary response 88 |
| chr6_+_32006159 | 0.37 |
ENST00000478281.1
ENST00000471671.1 ENST00000435122.2 |
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr9_+_100174232 | 0.36 |
ENST00000355295.4
|
TDRD7
|
tudor domain containing 7 |
| chr9_+_100174344 | 0.36 |
ENST00000422139.2
|
TDRD7
|
tudor domain containing 7 |
| chr11_-_60719213 | 0.35 |
ENST00000227880.3
|
SLC15A3
|
solute carrier family 15 (oligopeptide transporter), member 3 |
| chr10_+_13141441 | 0.35 |
ENST00000263036.5
|
OPTN
|
optineurin |
| chr5_-_176923803 | 0.34 |
ENST00000506161.1
|
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
| chr19_-_58609570 | 0.33 |
ENST00000600845.1
ENST00000240727.6 ENST00000600897.1 ENST00000421612.2 ENST00000601063.1 ENST00000601144.1 |
ZSCAN18
|
zinc finger and SCAN domain containing 18 |
| chr15_+_80351910 | 0.32 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr10_+_13141585 | 0.31 |
ENST00000378764.2
|
OPTN
|
optineurin |
| chr7_-_99774945 | 0.31 |
ENST00000292377.2
|
GPC2
|
glypican 2 |
| chr5_-_176924562 | 0.31 |
ENST00000359895.2
ENST00000355572.2 ENST00000355841.2 ENST00000393551.1 ENST00000505074.1 ENST00000356618.4 ENST00000393546.4 |
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
| chr17_-_39183452 | 0.31 |
ENST00000361883.5
|
KRTAP1-5
|
keratin associated protein 1-5 |
| chr1_-_16400086 | 0.30 |
ENST00000375662.4
|
FAM131C
|
family with sequence similarity 131, member C |
| chr16_-_88717482 | 0.30 |
ENST00000261623.3
|
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr21_-_45196326 | 0.30 |
ENST00000291568.5
|
CSTB
|
cystatin B (stefin B) |
| chr12_-_132834281 | 0.29 |
ENST00000411988.2
ENST00000535228.1 |
GALNT9
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 9 (GalNAc-T9) |
| chr12_+_132312931 | 0.27 |
ENST00000360564.1
ENST00000545671.1 ENST00000545790.1 |
MMP17
|
matrix metallopeptidase 17 (membrane-inserted) |
| chr1_-_153521714 | 0.27 |
ENST00000368713.3
|
S100A3
|
S100 calcium binding protein A3 |
| chr19_+_13906250 | 0.27 |
ENST00000254323.2
|
ZSWIM4
|
zinc finger, SWIM-type containing 4 |
| chr22_+_43506747 | 0.26 |
ENST00000216115.2
|
BIK
|
BCL2-interacting killer (apoptosis-inducing) |
| chr5_-_176923846 | 0.26 |
ENST00000506537.1
|
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
| chr16_+_29466426 | 0.26 |
ENST00000567248.1
|
SLX1B
|
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae) |
| chr17_-_79917645 | 0.25 |
ENST00000477214.1
|
NOTUM
|
notum pectinacetylesterase homolog (Drosophila) |
| chr10_+_13142075 | 0.25 |
ENST00000378757.2
ENST00000430081.1 ENST00000378752.3 ENST00000378748.3 |
OPTN
|
optineurin |
| chr11_-_65150103 | 0.24 |
ENST00000294187.6
ENST00000398802.1 ENST00000360662.3 ENST00000377152.2 ENST00000530936.1 |
SLC25A45
|
solute carrier family 25, member 45 |
| chr1_+_155293702 | 0.24 |
ENST00000368347.4
|
RUSC1
|
RUN and SH3 domain containing 1 |
| chr14_+_100848311 | 0.23 |
ENST00000542471.2
|
WDR25
|
WD repeat domain 25 |
| chr11_+_73675873 | 0.23 |
ENST00000537753.1
ENST00000542350.1 |
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr3_-_176914998 | 0.23 |
ENST00000431421.1
ENST00000422066.1 ENST00000413084.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr14_+_24641062 | 0.23 |
ENST00000311457.3
ENST00000557806.1 ENST00000559919.1 |
REC8
|
REC8 meiotic recombination protein |
| chr4_+_81951957 | 0.22 |
ENST00000282701.2
|
BMP3
|
bone morphogenetic protein 3 |
| chr16_+_56691838 | 0.22 |
ENST00000394501.2
|
MT1F
|
metallothionein 1F |
| chr19_+_45251804 | 0.22 |
ENST00000164227.5
|
BCL3
|
B-cell CLL/lymphoma 3 |
| chr2_+_85132749 | 0.22 |
ENST00000233143.4
|
TMSB10
|
thymosin beta 10 |
| chr19_-_10024496 | 0.22 |
ENST00000593091.1
|
OLFM2
|
olfactomedin 2 |
| chr1_-_21503337 | 0.22 |
ENST00000400422.1
ENST00000602326.1 ENST00000411888.1 ENST00000438975.1 |
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr19_-_1174226 | 0.22 |
ENST00000587024.1
ENST00000361757.3 |
SBNO2
|
strawberry notch homolog 2 (Drosophila) |
| chrX_+_51927919 | 0.21 |
ENST00000416960.1
|
MAGED4
|
melanoma antigen family D, 4 |
| chr16_-_29466285 | 0.21 |
ENST00000330978.3
|
BOLA2
|
bolA family member 2 |
| chrX_-_54070607 | 0.20 |
ENST00000338154.6
ENST00000338946.6 |
PHF8
|
PHD finger protein 8 |
| chr1_-_153521597 | 0.20 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chr10_+_13142225 | 0.19 |
ENST00000378747.3
|
OPTN
|
optineurin |
| chr3_-_129035120 | 0.19 |
ENST00000333762.4
|
H1FX
|
H1 histone family, member X |
| chr22_-_36635598 | 0.19 |
ENST00000454728.1
|
APOL2
|
apolipoprotein L, 2 |
| chr11_+_58938903 | 0.19 |
ENST00000532982.1
|
DTX4
|
deltex homolog 4 (Drosophila) |
| chr10_+_71561630 | 0.19 |
ENST00000398974.3
ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr2_+_101436564 | 0.19 |
ENST00000335681.5
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr21_+_37507210 | 0.18 |
ENST00000290354.5
|
CBR3
|
carbonyl reductase 3 |
| chr22_+_22723969 | 0.18 |
ENST00000390295.2
|
IGLV7-46
|
immunoglobulin lambda variable 7-46 (gene/pseudogene) |
| chr6_+_32006042 | 0.18 |
ENST00000418967.2
|
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr2_-_73869508 | 0.18 |
ENST00000272425.3
|
NAT8
|
N-acetyltransferase 8 (GCN5-related, putative) |
| chr1_-_6240183 | 0.18 |
ENST00000262450.3
ENST00000378021.1 |
CHD5
|
chromodomain helicase DNA binding protein 5 |
| chr20_-_56286479 | 0.18 |
ENST00000265626.4
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr16_-_30204987 | 0.18 |
ENST00000569282.1
ENST00000567436.1 |
BOLA2B
|
bolA family member 2B |
| chr19_+_589893 | 0.18 |
ENST00000251287.2
|
HCN2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2 |
| chr16_+_29465822 | 0.17 |
ENST00000330181.5
ENST00000351581.4 |
SLX1B
|
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae) |
| chr1_-_212873267 | 0.17 |
ENST00000243440.1
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr16_-_30205627 | 0.17 |
ENST00000305321.4
|
BOLA2B
|
bolA family member 2B |
| chr9_-_114246332 | 0.17 |
ENST00000602978.1
|
KIAA0368
|
KIAA0368 |
| chr15_+_90728145 | 0.17 |
ENST00000561085.1
ENST00000379122.3 ENST00000332496.6 |
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr17_-_42988004 | 0.17 |
ENST00000586125.1
ENST00000591880.1 |
GFAP
|
glial fibrillary acidic protein |
| chr11_+_69061594 | 0.17 |
ENST00000441339.2
ENST00000308946.3 ENST00000535407.1 |
MYEOV
|
myeloma overexpressed |
| chr3_-_50540854 | 0.16 |
ENST00000423994.2
ENST00000424201.2 ENST00000479441.1 ENST00000429770.1 |
CACNA2D2
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2 |
| chr16_+_30205225 | 0.16 |
ENST00000345535.4
ENST00000251303.6 |
SLX1A
|
SLX1 structure-specific endonuclease subunit homolog A (S. cerevisiae) |
| chr12_+_49932886 | 0.16 |
ENST00000257981.6
|
KCNH3
|
potassium voltage-gated channel, subfamily H (eag-related), member 3 |
| chr19_+_18077881 | 0.16 |
ENST00000609922.1
|
KCNN1
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 |
| chr16_+_56691911 | 0.15 |
ENST00000568475.1
|
MT1F
|
metallothionein 1F |
| chr16_+_56691606 | 0.15 |
ENST00000334350.6
|
MT1F
|
metallothionein 1F |
| chr16_+_9185450 | 0.15 |
ENST00000327827.7
|
C16orf72
|
chromosome 16 open reading frame 72 |
| chr3_+_11196206 | 0.15 |
ENST00000431010.2
|
HRH1
|
histamine receptor H1 |
| chr1_+_28995231 | 0.15 |
ENST00000373816.1
|
GMEB1
|
glucocorticoid modulatory element binding protein 1 |
| chr16_+_4897632 | 0.15 |
ENST00000262376.6
|
UBN1
|
ubinuclein 1 |
| chr2_-_233792837 | 0.14 |
ENST00000373552.4
ENST00000409079.1 |
NGEF
|
neuronal guanine nucleotide exchange factor |
| chrX_-_54070388 | 0.14 |
ENST00000415025.1
|
PHF8
|
PHD finger protein 8 |
| chr17_+_78194205 | 0.14 |
ENST00000573809.1
ENST00000361193.3 ENST00000574967.1 ENST00000576126.1 ENST00000411502.3 ENST00000546047.2 |
SLC26A11
|
solute carrier family 26 (anion exchanger), member 11 |
| chr19_+_17970693 | 0.14 |
ENST00000600147.1
ENST00000599898.1 |
RPL18A
|
ribosomal protein L18a |
| chr5_-_157002775 | 0.14 |
ENST00000257527.4
|
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr12_-_7244469 | 0.14 |
ENST00000538050.1
ENST00000536053.2 |
C1R
|
complement component 1, r subcomponent |
| chr16_-_2059797 | 0.14 |
ENST00000563630.1
|
ZNF598
|
zinc finger protein 598 |
| chr16_+_58035277 | 0.14 |
ENST00000219281.3
ENST00000539737.2 ENST00000423271.3 ENST00000561568.1 ENST00000563149.1 |
USB1
|
U6 snRNA biogenesis 1 |
| chr6_-_38607673 | 0.14 |
ENST00000481247.1
|
BTBD9
|
BTB (POZ) domain containing 9 |
| chr10_+_112257596 | 0.14 |
ENST00000369583.3
|
DUSP5
|
dual specificity phosphatase 5 |
| chr19_-_23869999 | 0.13 |
ENST00000601935.1
ENST00000359788.4 ENST00000600313.1 ENST00000596211.1 ENST00000599168.1 |
ZNF675
|
zinc finger protein 675 |
| chr12_-_51420128 | 0.13 |
ENST00000262051.7
ENST00000547732.1 ENST00000262052.5 ENST00000546488.1 ENST00000550714.1 ENST00000548193.1 ENST00000547579.1 ENST00000546743.1 |
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr11_+_64781575 | 0.13 |
ENST00000246747.4
ENST00000529384.1 |
ARL2
|
ADP-ribosylation factor-like 2 |
| chr13_-_41111323 | 0.13 |
ENST00000595486.1
|
AL133318.1
|
Uncharacterized protein |
| chr11_-_125932685 | 0.13 |
ENST00000527967.1
|
CDON
|
cell adhesion associated, oncogene regulated |
| chr22_+_19705928 | 0.13 |
ENST00000383045.3
ENST00000438754.2 |
SEPT5
|
septin 5 |
| chr14_+_24630465 | 0.13 |
ENST00000557894.1
ENST00000559284.1 ENST00000560275.1 |
IRF9
|
interferon regulatory factor 9 |
| chr19_+_17970677 | 0.13 |
ENST00000222247.5
|
RPL18A
|
ribosomal protein L18a |
| chr5_-_157002749 | 0.13 |
ENST00000517905.1
ENST00000430702.2 ENST00000394020.1 |
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr1_+_3370990 | 0.13 |
ENST00000378378.4
|
ARHGEF16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr17_-_56065540 | 0.13 |
ENST00000583932.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr17_+_5981270 | 0.13 |
ENST00000571973.1
|
WSCD1
|
WSC domain containing 1 |
| chr15_+_80351977 | 0.13 |
ENST00000559157.1
ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr17_-_78194124 | 0.12 |
ENST00000570427.1
ENST00000570923.1 |
SGSH
|
N-sulfoglucosamine sulfohydrolase |
| chr4_-_15656923 | 0.12 |
ENST00000382358.4
ENST00000412094.2 ENST00000341285.3 |
FBXL5
|
F-box and leucine-rich repeat protein 5 |
| chr2_+_23608064 | 0.12 |
ENST00000486442.1
|
KLHL29
|
kelch-like family member 29 |
| chr22_-_20368028 | 0.12 |
ENST00000404912.1
|
GGTLC3
|
gamma-glutamyltransferase light chain 3 |
| chr11_+_134146635 | 0.12 |
ENST00000431683.2
|
GLB1L3
|
galactosidase, beta 1-like 3 |
| chr17_+_48638371 | 0.12 |
ENST00000360761.4
ENST00000352832.5 ENST00000354983.4 |
CACNA1G
|
calcium channel, voltage-dependent, T type, alpha 1G subunit |
| chr5_+_43192311 | 0.12 |
ENST00000326035.2
|
NIM1
|
NIM1 serine/threonine protein kinase |
| chr16_+_2059872 | 0.12 |
ENST00000567649.1
|
NPW
|
neuropeptide W |
| chr6_+_36098262 | 0.12 |
ENST00000373761.6
ENST00000373766.5 |
MAPK13
|
mitogen-activated protein kinase 13 |
| chr15_-_41408409 | 0.12 |
ENST00000361937.3
|
INO80
|
INO80 complex subunit |
| chr1_-_235814048 | 0.12 |
ENST00000450593.1
ENST00000366598.4 |
GNG4
|
guanine nucleotide binding protein (G protein), gamma 4 |
| chr3_-_134093395 | 0.12 |
ENST00000249883.5
|
AMOTL2
|
angiomotin like 2 |
| chr6_+_36097992 | 0.12 |
ENST00000211287.4
|
MAPK13
|
mitogen-activated protein kinase 13 |
| chr16_+_24550857 | 0.12 |
ENST00000568015.1
|
RBBP6
|
retinoblastoma binding protein 6 |
| chr5_-_126366500 | 0.11 |
ENST00000308660.5
|
MARCH3
|
membrane-associated ring finger (C3HC4) 3, E3 ubiquitin protein ligase |
| chr11_+_66790816 | 0.11 |
ENST00000527043.1
|
SYT12
|
synaptotagmin XII |
| chr3_+_183948161 | 0.11 |
ENST00000426955.2
|
VWA5B2
|
von Willebrand factor A domain containing 5B2 |
| chr17_-_39646116 | 0.11 |
ENST00000328119.6
|
KRT36
|
keratin 36 |
| chr11_+_120081475 | 0.11 |
ENST00000328965.4
|
OAF
|
OAF homolog (Drosophila) |
| chr5_-_131563501 | 0.11 |
ENST00000401867.1
ENST00000379086.1 ENST00000418055.1 ENST00000453286.1 ENST00000166534.4 |
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chr10_-_126849626 | 0.11 |
ENST00000530884.1
|
CTBP2
|
C-terminal binding protein 2 |
| chr2_+_109150887 | 0.11 |
ENST00000428064.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr6_+_36098363 | 0.11 |
ENST00000373759.1
|
MAPK13
|
mitogen-activated protein kinase 13 |
| chr22_-_39636914 | 0.11 |
ENST00000381551.4
|
PDGFB
|
platelet-derived growth factor beta polypeptide |
| chr6_+_15249128 | 0.11 |
ENST00000397311.3
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr2_+_105050794 | 0.11 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr12_-_51420108 | 0.11 |
ENST00000547198.1
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr18_-_51750948 | 0.11 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr19_+_56165480 | 0.11 |
ENST00000450554.2
|
U2AF2
|
U2 small nuclear RNA auxiliary factor 2 |
| chr16_-_29465668 | 0.11 |
ENST00000569622.1
|
RP11-345J4.5
|
BolA-like protein 2 |
| chr14_+_29236269 | 0.11 |
ENST00000313071.4
|
FOXG1
|
forkhead box G1 |
| chrX_+_134555863 | 0.10 |
ENST00000417443.2
|
LINC00086
|
long intergenic non-protein coding RNA 86 |
| chr1_+_43232913 | 0.10 |
ENST00000372525.5
ENST00000536543.1 |
C1orf50
|
chromosome 1 open reading frame 50 |
| chr19_-_40791211 | 0.10 |
ENST00000579047.1
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr11_-_65667997 | 0.10 |
ENST00000312562.2
ENST00000534222.1 |
FOSL1
|
FOS-like antigen 1 |
| chr16_+_89989687 | 0.10 |
ENST00000315491.7
ENST00000555576.1 ENST00000554336.1 ENST00000553967.1 |
TUBB3
|
Tubulin beta-3 chain |
| chr2_+_219824357 | 0.10 |
ENST00000302625.4
|
CDK5R2
|
cyclin-dependent kinase 5, regulatory subunit 2 (p39) |
| chr5_-_110074603 | 0.10 |
ENST00000515278.2
|
TMEM232
|
transmembrane protein 232 |
| chr16_+_777739 | 0.10 |
ENST00000563792.1
|
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr11_-_67276100 | 0.10 |
ENST00000301488.3
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2 |
| chrX_+_13707235 | 0.10 |
ENST00000464506.1
|
RAB9A
|
RAB9A, member RAS oncogene family |
| chr20_+_44563267 | 0.10 |
ENST00000372409.3
|
PCIF1
|
PDX1 C-terminal inhibiting factor 1 |
| chr2_-_202645612 | 0.10 |
ENST00000409632.2
ENST00000410052.1 ENST00000467448.1 |
ALS2
|
amyotrophic lateral sclerosis 2 (juvenile) |
| chr11_-_65667884 | 0.10 |
ENST00000448083.2
ENST00000531493.1 ENST00000532401.1 |
FOSL1
|
FOS-like antigen 1 |
| chr3_-_118753792 | 0.10 |
ENST00000480431.1
|
IGSF11
|
immunoglobulin superfamily, member 11 |
| chr7_-_150020578 | 0.10 |
ENST00000478393.1
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast) |
| chr5_+_176873446 | 0.10 |
ENST00000507881.1
|
PRR7
|
proline rich 7 (synaptic) |
| chr17_+_77030267 | 0.10 |
ENST00000581774.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr11_-_407103 | 0.10 |
ENST00000526395.1
|
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr15_-_91565770 | 0.10 |
ENST00000535906.1
ENST00000333371.3 |
VPS33B
|
vacuolar protein sorting 33 homolog B (yeast) |
| chr2_+_10183651 | 0.10 |
ENST00000305883.1
|
KLF11
|
Kruppel-like factor 11 |
| chr1_+_6673745 | 0.09 |
ENST00000377648.4
|
PHF13
|
PHD finger protein 13 |
| chr9_-_74383799 | 0.09 |
ENST00000377044.4
|
TMEM2
|
transmembrane protein 2 |
| chr17_-_58469474 | 0.09 |
ENST00000300896.4
|
USP32
|
ubiquitin specific peptidase 32 |
| chr1_+_2407754 | 0.09 |
ENST00000419816.2
ENST00000378486.3 ENST00000378488.3 ENST00000288766.5 |
PLCH2
|
phospholipase C, eta 2 |
| chr1_+_156024552 | 0.09 |
ENST00000368304.5
ENST00000368302.3 |
LAMTOR2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr19_-_2721412 | 0.09 |
ENST00000323469.4
|
DIRAS1
|
DIRAS family, GTP-binding RAS-like 1 |
| chr10_+_71561649 | 0.09 |
ENST00000398978.3
ENST00000354547.3 ENST00000357811.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr12_-_48152853 | 0.09 |
ENST00000171000.4
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr1_+_183441500 | 0.09 |
ENST00000456731.2
|
SMG7
|
SMG7 nonsense mediated mRNA decay factor |
| chr19_+_984313 | 0.09 |
ENST00000251289.5
ENST00000587001.2 ENST00000607440.1 |
WDR18
|
WD repeat domain 18 |
| chr11_-_71159458 | 0.09 |
ENST00000355527.3
|
DHCR7
|
7-dehydrocholesterol reductase |
| chr15_-_41408339 | 0.09 |
ENST00000401393.3
|
INO80
|
INO80 complex subunit |
| chr16_-_8962853 | 0.09 |
ENST00000565287.1
ENST00000311052.5 |
CARHSP1
|
calcium regulated heat stable protein 1, 24kDa |
| chr17_-_15902951 | 0.09 |
ENST00000472495.1
|
ZSWIM7
|
zinc finger, SWIM-type containing 7 |
| chr19_+_47759716 | 0.09 |
ENST00000221922.6
|
CCDC9
|
coiled-coil domain containing 9 |
| chr17_-_1083078 | 0.09 |
ENST00000574266.1
ENST00000302538.5 |
ABR
|
active BCR-related |
| chr15_-_91565743 | 0.09 |
ENST00000535843.1
|
VPS33B
|
vacuolar protein sorting 33 homolog B (yeast) |
| chr17_-_76124812 | 0.09 |
ENST00000592063.1
ENST00000589271.1 ENST00000322933.4 ENST00000589553.1 |
TMC6
|
transmembrane channel-like 6 |
| chr10_-_23633720 | 0.09 |
ENST00000323327.4
|
C10orf67
|
chromosome 10 open reading frame 67 |
| chr17_-_80291818 | 0.09 |
ENST00000269389.3
ENST00000581691.1 |
SECTM1
|
secreted and transmembrane 1 |
| chr2_+_242255573 | 0.08 |
ENST00000420786.1
|
SEPT2
|
septin 2 |
| chr22_-_31328881 | 0.08 |
ENST00000445980.1
|
MORC2
|
MORC family CW-type zinc finger 2 |
| chr17_+_45331184 | 0.08 |
ENST00000559488.1
ENST00000571680.1 ENST00000435993.2 |
ITGB3
|
integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) |
| chr6_-_85473156 | 0.08 |
ENST00000606784.1
ENST00000606325.1 |
TBX18
|
T-box 18 |
| chr11_-_34379546 | 0.08 |
ENST00000435224.2
|
ABTB2
|
ankyrin repeat and BTB (POZ) domain containing 2 |
| chr16_+_4897912 | 0.08 |
ENST00000545171.1
|
UBN1
|
ubinuclein 1 |
| chr16_+_30205754 | 0.08 |
ENST00000354723.6
ENST00000355544.5 |
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr2_+_79740118 | 0.08 |
ENST00000496558.1
ENST00000451966.1 |
CTNNA2
|
catenin (cadherin-associated protein), alpha 2 |
| chr14_+_103243813 | 0.08 |
ENST00000560371.1
ENST00000347662.4 ENST00000392745.2 ENST00000539721.1 ENST00000560463.1 |
TRAF3
|
TNF receptor-associated factor 3 |
| chr11_+_12696102 | 0.08 |
ENST00000527636.1
ENST00000527376.1 |
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr16_+_56623433 | 0.08 |
ENST00000570176.1
|
MT3
|
metallothionein 3 |
| chr7_-_100808394 | 0.08 |
ENST00000445482.2
|
VGF
|
VGF nerve growth factor inducible |
| chr19_+_48949087 | 0.08 |
ENST00000598711.1
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr19_-_18385246 | 0.08 |
ENST00000608950.1
ENST00000600328.3 ENST00000600359.3 ENST00000392413.4 |
KIAA1683
|
KIAA1683 |
| chr6_+_123317116 | 0.08 |
ENST00000275162.5
|
CLVS2
|
clavesin 2 |
| chr16_+_2039946 | 0.08 |
ENST00000248121.2
ENST00000568896.1 |
SYNGR3
|
synaptogyrin 3 |
| chr16_-_1993260 | 0.08 |
ENST00000361871.3
|
MSRB1
|
methionine sulfoxide reductase B1 |
| chr19_-_48753028 | 0.08 |
ENST00000522431.1
|
CARD8
|
caspase recruitment domain family, member 8 |
| chr9_+_132565418 | 0.08 |
ENST00000259339.2
|
TOR1B
|
torsin family 1, member B (torsin B) |
| chr7_-_150020750 | 0.08 |
ENST00000539352.1
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast) |
| chr7_+_75511362 | 0.08 |
ENST00000428119.1
|
RHBDD2
|
rhomboid domain containing 2 |
| chr5_-_131563474 | 0.08 |
ENST00000417528.1
|
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chr1_-_11751529 | 0.08 |
ENST00000376672.1
|
MAD2L2
|
MAD2 mitotic arrest deficient-like 2 (yeast) |
| chr14_+_64971438 | 0.08 |
ENST00000555321.1
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZIC3_ZIC4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0061743 | motor learning(GO:0061743) maintenance of synapse structure(GO:0099558) |
| 0.2 | 0.5 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.2 | 1.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.7 | GO:0010847 | regulation of chromatin assembly(GO:0010847) protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 0.4 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 0.4 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.1 | 0.3 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.1 | 0.3 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.1 | 0.7 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 0.2 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.1 | 0.3 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 0.3 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.1 | 0.2 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.2 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.3 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.2 | GO:0044571 | [2Fe-2S] cluster assembly(GO:0044571) |
| 0.0 | 0.5 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.0 | 0.1 | GO:0061184 | dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) positive regulation of dermatome development(GO:0061184) |
| 0.0 | 0.2 | GO:0042376 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.0 | 0.1 | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning(GO:0021919) |
| 0.0 | 0.1 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.0 | 0.2 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.5 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.0 | 0.2 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.1 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.2 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.0 | 0.1 | GO:0097212 | cadmium ion homeostasis(GO:0055073) lysosomal membrane organization(GO:0097212) negative regulation of hydrogen peroxide catabolic process(GO:2000296) regulation of oxygen metabolic process(GO:2000374) |
| 0.0 | 0.1 | GO:0090678 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.0 | 0.2 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.3 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.0 | 0.1 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 0.0 | 0.6 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.2 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.0 | 0.1 | GO:0030822 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.0 | 0.4 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.1 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.2 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.8 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.3 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 | 0.1 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.1 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.0 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.0 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.1 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0071224 | cellular response to peptidoglycan(GO:0071224) |
| 0.0 | 0.1 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.2 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.5 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.2 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.0 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 | 0.1 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.2 | GO:0070305 | response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.0 | 0.0 | GO:0044793 | negative regulation by host of viral process(GO:0044793) negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.0 | GO:0000472 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.0 | 0.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.2 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.1 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.2 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.1 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.1 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.2 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.1 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.1 | GO:0097473 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.0 | 0.1 | GO:0043405 | regulation of MAP kinase activity(GO:0043405) positive regulation of MAP kinase activity(GO:0043406) |
| 0.0 | 0.0 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.2 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.4 | GO:0072663 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.0 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.0 | GO:0060492 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) positive regulation of branching involved in lung morphogenesis(GO:0061047) |
| 0.0 | 0.0 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.1 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.0 | 0.2 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.1 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.0 | GO:0015882 | L-ascorbic acid transport(GO:0015882) molecular hydrogen transport(GO:0015993) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.1 | 0.5 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.1 | 0.3 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.1 | 0.2 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.3 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.7 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.2 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.2 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.1 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0097196 | Shu complex(GO:0097196) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.3 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.0 | 0.0 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.1 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.0 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.0 | 0.0 | GO:0070703 | inner mucus layer(GO:0070702) outer mucus layer(GO:0070703) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.2 | GO:0071439 | clathrin complex(GO:0071439) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 0.3 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.1 | 0.2 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 0.5 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 0.3 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.2 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.2 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 1.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.3 | GO:0015639 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.1 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.2 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.0 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.0 | 0.1 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.2 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.6 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.0 | GO:0036328 | VEGF-C-activated receptor activity(GO:0036328) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.0 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.1 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.0 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.2 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.5 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.0 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.8 | PID AURORA A PATHWAY | Aurora A signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |