Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for ZIC2_GLI1
Z-value: 0.61
Transcription factors associated with ZIC2_GLI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
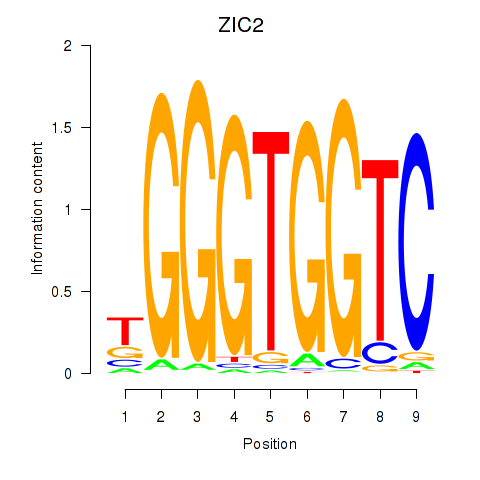
ZIC2
|
ENSG00000043355.6 | Zic family member 2 |
|
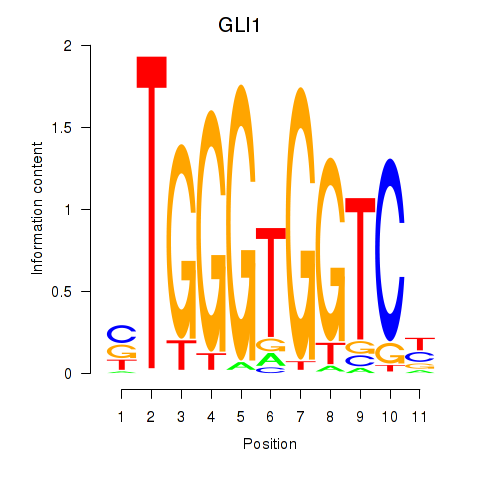
GLI1
|
ENSG00000111087.5 | GLI family zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZIC2 | hg19_v2_chr13_+_100634004_100634026 | -0.95 | 5.2e-02 | Click! |
| GLI1 | hg19_v2_chr12_+_57857475_57857475 | -0.67 | 3.3e-01 | Click! |
Activity profile of ZIC2_GLI1 motif
Sorted Z-values of ZIC2_GLI1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_26027480 | 0.71 |
ENST00000377364.3
|
HIST1H4B
|
histone cluster 1, H4b |
| chr22_-_50970566 | 0.58 |
ENST00000405135.1
ENST00000401779.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr6_-_32821599 | 0.56 |
ENST00000354258.4
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr22_-_50970506 | 0.51 |
ENST00000428989.2
ENST00000403326.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr19_-_55881741 | 0.47 |
ENST00000264563.2
ENST00000590625.1 ENST00000585513.1 |
IL11
|
interleukin 11 |
| chr5_-_168727786 | 0.47 |
ENST00000332966.8
|
SLIT3
|
slit homolog 3 (Drosophila) |
| chr22_-_50970919 | 0.43 |
ENST00000329363.4
ENST00000437588.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr17_-_15496722 | 0.39 |
ENST00000472534.1
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr2_+_149402553 | 0.36 |
ENST00000258484.6
ENST00000409654.1 |
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr14_+_101292445 | 0.35 |
ENST00000429159.2
ENST00000520714.1 ENST00000522771.2 ENST00000424076.3 ENST00000423456.1 ENST00000521404.1 ENST00000556736.1 ENST00000451743.2 ENST00000398518.2 ENST00000554639.1 ENST00000452120.2 ENST00000519709.1 ENST00000412736.2 |
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr19_+_676385 | 0.31 |
ENST00000166139.4
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein) |
| chr17_+_40834580 | 0.30 |
ENST00000264638.4
|
CNTNAP1
|
contactin associated protein 1 |
| chr14_-_24584138 | 0.30 |
ENST00000558280.1
ENST00000561028.1 |
NRL
|
neural retina leucine zipper |
| chr22_-_46373004 | 0.29 |
ENST00000339464.4
|
WNT7B
|
wingless-type MMTV integration site family, member 7B |
| chr1_+_183155373 | 0.29 |
ENST00000493293.1
ENST00000264144.4 |
LAMC2
|
laminin, gamma 2 |
| chr17_-_34122596 | 0.27 |
ENST00000250144.8
|
MMP28
|
matrix metallopeptidase 28 |
| chr5_-_141704566 | 0.27 |
ENST00000344120.4
ENST00000434127.2 |
SPRY4
|
sprouty homolog 4 (Drosophila) |
| chr7_+_114562616 | 0.26 |
ENST00000448022.1
|
MDFIC
|
MyoD family inhibitor domain containing |
| chr7_-_106301405 | 0.25 |
ENST00000523505.1
|
CCDC71L
|
coiled-coil domain containing 71-like |
| chrX_+_51928002 | 0.24 |
ENST00000375626.3
|
MAGED4
|
melanoma antigen family D, 4 |
| chr14_-_61124977 | 0.23 |
ENST00000554986.1
|
SIX1
|
SIX homeobox 1 |
| chr8_+_94752349 | 0.23 |
ENST00000391680.1
|
RBM12B-AS1
|
RBM12B antisense RNA 1 |
| chr5_-_150460539 | 0.22 |
ENST00000520931.1
ENST00000520695.1 ENST00000521591.1 ENST00000518977.1 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr17_-_40835076 | 0.22 |
ENST00000591765.1
|
CCR10
|
chemokine (C-C motif) receptor 10 |
| chr20_+_43343886 | 0.21 |
ENST00000190983.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr5_-_168727713 | 0.21 |
ENST00000404867.3
|
SLIT3
|
slit homolog 3 (Drosophila) |
| chr6_-_32784687 | 0.21 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr20_+_43343517 | 0.21 |
ENST00000372865.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr7_-_92465868 | 0.21 |
ENST00000424848.2
|
CDK6
|
cyclin-dependent kinase 6 |
| chr18_+_21529811 | 0.21 |
ENST00000588004.1
|
LAMA3
|
laminin, alpha 3 |
| chr16_-_1031259 | 0.20 |
ENST00000563837.1
ENST00000563863.1 ENST00000565069.1 ENST00000570014.1 |
RP11-161M6.2
LMF1
|
RP11-161M6.2 lipase maturation factor 1 |
| chr18_-_44775554 | 0.20 |
ENST00000425639.1
ENST00000400404.1 |
SKOR2
|
SKI family transcriptional corepressor 2 |
| chr17_-_37557846 | 0.20 |
ENST00000394294.3
ENST00000583610.1 ENST00000264658.6 |
FBXL20
|
F-box and leucine-rich repeat protein 20 |
| chr10_-_18948208 | 0.20 |
ENST00000607346.1
|
ARL5B-AS1
|
ARL5B antisense RNA 1 |
| chr9_-_98079965 | 0.19 |
ENST00000289081.3
|
FANCC
|
Fanconi anemia, complementation group C |
| chr20_+_43343476 | 0.19 |
ENST00000372868.2
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr17_-_27278445 | 0.19 |
ENST00000268756.3
ENST00000584685.1 |
PHF12
|
PHD finger protein 12 |
| chr14_+_24584508 | 0.18 |
ENST00000559354.1
ENST00000560459.1 ENST00000559593.1 ENST00000396941.4 ENST00000396936.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr7_-_108097144 | 0.18 |
ENST00000418239.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr12_-_6798025 | 0.18 |
ENST00000542351.1
ENST00000538829.1 |
ZNF384
|
zinc finger protein 384 |
| chr5_-_150460914 | 0.17 |
ENST00000389378.2
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chrX_+_47444613 | 0.17 |
ENST00000445623.1
|
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr15_+_67430339 | 0.17 |
ENST00000439724.3
|
SMAD3
|
SMAD family member 3 |
| chr16_+_86612112 | 0.17 |
ENST00000320241.3
|
FOXL1
|
forkhead box L1 |
| chr17_+_26989109 | 0.17 |
ENST00000314616.6
ENST00000347486.4 |
SUPT6H
|
suppressor of Ty 6 homolog (S. cerevisiae) |
| chr7_-_100034060 | 0.17 |
ENST00000292330.2
|
PPP1R35
|
protein phosphatase 1, regulatory subunit 35 |
| chr16_+_57844549 | 0.17 |
ENST00000564282.1
|
CTD-2600O9.1
|
uncharacterized protein LOC388282 |
| chrX_+_48687283 | 0.17 |
ENST00000338270.1
|
ERAS
|
ES cell expressed Ras |
| chr17_-_37558776 | 0.16 |
ENST00000577399.1
|
FBXL20
|
F-box and leucine-rich repeat protein 20 |
| chr10_-_97050777 | 0.16 |
ENST00000329399.6
|
PDLIM1
|
PDZ and LIM domain 1 |
| chr12_-_6798616 | 0.16 |
ENST00000355772.4
ENST00000417772.3 ENST00000396801.3 ENST00000396799.2 |
ZNF384
|
zinc finger protein 384 |
| chr9_+_135037334 | 0.16 |
ENST00000393229.3
ENST00000360670.3 ENST00000393228.4 ENST00000372179.3 |
NTNG2
|
netrin G2 |
| chr1_-_12677714 | 0.15 |
ENST00000376223.2
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr20_+_62327996 | 0.15 |
ENST00000369996.1
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy |
| chr20_-_30310656 | 0.15 |
ENST00000376055.4
|
BCL2L1
|
BCL2-like 1 |
| chr7_+_119913688 | 0.15 |
ENST00000331113.4
|
KCND2
|
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr20_-_30310693 | 0.14 |
ENST00000307677.4
ENST00000420653.1 |
BCL2L1
|
BCL2-like 1 |
| chr3_-_88108212 | 0.14 |
ENST00000482016.1
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr6_-_29600832 | 0.14 |
ENST00000377016.4
ENST00000376977.3 ENST00000377034.4 |
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr2_-_133427767 | 0.14 |
ENST00000397463.2
|
LYPD1
|
LY6/PLAUR domain containing 1 |
| chr19_+_4198072 | 0.14 |
ENST00000262970.5
|
ANKRD24
|
ankyrin repeat domain 24 |
| chr1_+_36621529 | 0.14 |
ENST00000316156.4
|
MAP7D1
|
MAP7 domain containing 1 |
| chr3_+_47844615 | 0.14 |
ENST00000348968.4
|
DHX30
|
DEAH (Asp-Glu-Ala-His) box helicase 30 |
| chr7_+_114562909 | 0.14 |
ENST00000423503.1
ENST00000427207.1 |
MDFIC
|
MyoD family inhibitor domain containing |
| chr3_+_47844399 | 0.13 |
ENST00000446256.2
ENST00000445061.1 |
DHX30
|
DEAH (Asp-Glu-Ala-His) box helicase 30 |
| chr17_+_73089382 | 0.13 |
ENST00000538213.2
ENST00000584118.1 |
SLC16A5
|
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr1_-_205782304 | 0.13 |
ENST00000367137.3
|
SLC41A1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr14_+_64971438 | 0.13 |
ENST00000555321.1
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr10_-_77161533 | 0.13 |
ENST00000535216.1
|
ZNF503
|
zinc finger protein 503 |
| chr6_-_19804973 | 0.13 |
ENST00000457670.1
ENST00000607810.1 ENST00000606628.1 |
RP4-625H18.2
|
RP4-625H18.2 |
| chr12_-_45270077 | 0.13 |
ENST00000551601.1
ENST00000549027.1 ENST00000452445.2 |
NELL2
|
NEL-like 2 (chicken) |
| chr6_-_31620403 | 0.13 |
ENST00000451898.1
ENST00000439687.2 ENST00000362049.6 ENST00000424480.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr12_-_6798523 | 0.13 |
ENST00000319770.3
|
ZNF384
|
zinc finger protein 384 |
| chr14_-_51561784 | 0.13 |
ENST00000360392.4
|
TRIM9
|
tripartite motif containing 9 |
| chr7_-_112726393 | 0.13 |
ENST00000449591.1
ENST00000449735.1 ENST00000438062.1 ENST00000424100.1 |
GPR85
|
G protein-coupled receptor 85 |
| chr17_-_7137582 | 0.12 |
ENST00000575756.1
ENST00000575458.1 |
DVL2
|
dishevelled segment polarity protein 2 |
| chr7_+_149411860 | 0.12 |
ENST00000486744.1
|
KRBA1
|
KRAB-A domain containing 1 |
| chr1_+_24120143 | 0.12 |
ENST00000374501.1
|
LYPLA2
|
lysophospholipase II |
| chr14_+_92980111 | 0.12 |
ENST00000216487.7
ENST00000557762.1 |
RIN3
|
Ras and Rab interactor 3 |
| chr15_-_41408409 | 0.12 |
ENST00000361937.3
|
INO80
|
INO80 complex subunit |
| chr4_-_134070250 | 0.12 |
ENST00000505289.1
ENST00000509715.1 |
RP11-9G1.3
|
RP11-9G1.3 |
| chr15_-_83224682 | 0.12 |
ENST00000562833.1
|
RP11-152F13.10
|
RP11-152F13.10 |
| chr6_-_31620455 | 0.12 |
ENST00000437771.1
ENST00000404765.2 ENST00000375964.6 ENST00000211379.5 |
BAG6
|
BCL2-associated athanogene 6 |
| chr6_-_19804877 | 0.11 |
ENST00000447250.1
|
RP4-625H18.2
|
RP4-625H18.2 |
| chr7_+_116312411 | 0.11 |
ENST00000456159.1
ENST00000397752.3 ENST00000318493.6 |
MET
|
met proto-oncogene |
| chr12_+_57522439 | 0.11 |
ENST00000338962.4
|
LRP1
|
low density lipoprotein receptor-related protein 1 |
| chr13_-_30880979 | 0.11 |
ENST00000414289.1
|
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr14_+_24583836 | 0.11 |
ENST00000559115.1
ENST00000558215.1 ENST00000557810.1 ENST00000561375.1 ENST00000446197.3 ENST00000559796.1 ENST00000560713.1 ENST00000560901.1 ENST00000559382.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr20_-_30310797 | 0.11 |
ENST00000422920.1
|
BCL2L1
|
BCL2-like 1 |
| chr21_-_28215332 | 0.11 |
ENST00000517777.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr18_-_31803435 | 0.11 |
ENST00000589544.1
ENST00000269185.4 ENST00000261592.5 |
NOL4
|
nucleolar protein 4 |
| chrX_-_37706661 | 0.11 |
ENST00000432389.2
ENST00000378581.3 |
DYNLT3
|
dynein, light chain, Tctex-type 3 |
| chr6_+_17281802 | 0.10 |
ENST00000509686.1
|
RBM24
|
RNA binding motif protein 24 |
| chr17_+_4843654 | 0.10 |
ENST00000575111.1
|
RNF167
|
ring finger protein 167 |
| chr16_+_86600857 | 0.10 |
ENST00000320354.4
|
FOXC2
|
forkhead box C2 (MFH-1, mesenchyme forkhead 1) |
| chr4_-_149363376 | 0.10 |
ENST00000512865.1
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr11_-_65321198 | 0.10 |
ENST00000530426.1
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr6_-_31620095 | 0.10 |
ENST00000424176.1
ENST00000456622.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr10_-_77161650 | 0.10 |
ENST00000372524.4
|
ZNF503
|
zinc finger protein 503 |
| chr6_-_31620149 | 0.10 |
ENST00000435080.1
ENST00000375976.4 ENST00000441054.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr19_-_13617037 | 0.10 |
ENST00000360228.5
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr6_-_29600559 | 0.10 |
ENST00000476670.1
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr15_+_60296421 | 0.10 |
ENST00000396057.4
|
FOXB1
|
forkhead box B1 |
| chrX_+_70364667 | 0.10 |
ENST00000536169.1
ENST00000395855.2 ENST00000374051.3 ENST00000358741.3 |
NLGN3
|
neuroligin 3 |
| chr2_+_28113583 | 0.10 |
ENST00000344773.2
ENST00000379624.1 ENST00000342045.2 ENST00000379632.2 ENST00000361704.2 |
BRE
|
brain and reproductive organ-expressed (TNFRSF1A modulator) |
| chr19_-_44285401 | 0.09 |
ENST00000262888.3
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr21_-_36259445 | 0.09 |
ENST00000399240.1
|
RUNX1
|
runt-related transcription factor 1 |
| chr1_+_155293702 | 0.09 |
ENST00000368347.4
|
RUSC1
|
RUN and SH3 domain containing 1 |
| chr17_+_17876127 | 0.09 |
ENST00000582416.1
ENST00000313838.8 ENST00000411504.2 ENST00000581264.1 ENST00000399187.1 ENST00000479684.2 ENST00000584166.1 ENST00000585108.1 ENST00000399182.1 ENST00000579977.1 |
LRRC48
|
leucine rich repeat containing 48 |
| chr17_-_76732928 | 0.09 |
ENST00000589768.1
|
CYTH1
|
cytohesin 1 |
| chr11_-_57103327 | 0.09 |
ENST00000529002.1
ENST00000278412.2 |
SSRP1
|
structure specific recognition protein 1 |
| chr11_-_66104237 | 0.09 |
ENST00000530056.1
|
RIN1
|
Ras and Rab interactor 1 |
| chr15_-_41408339 | 0.09 |
ENST00000401393.3
|
INO80
|
INO80 complex subunit |
| chr11_-_46848393 | 0.09 |
ENST00000526496.1
|
CKAP5
|
cytoskeleton associated protein 5 |
| chr8_+_31497271 | 0.09 |
ENST00000520407.1
|
NRG1
|
neuregulin 1 |
| chr10_-_77161004 | 0.09 |
ENST00000418818.2
|
RP11-399K21.11
|
RP11-399K21.11 |
| chrX_-_51812268 | 0.09 |
ENST00000486010.1
ENST00000497164.1 ENST00000360134.6 ENST00000485287.1 ENST00000335504.5 ENST00000431659.1 |
MAGED4B
|
melanoma antigen family D, 4B |
| chr2_-_213403565 | 0.09 |
ENST00000342788.4
ENST00000436443.1 |
ERBB4
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 4 |
| chr17_+_4843679 | 0.09 |
ENST00000576229.1
|
RNF167
|
ring finger protein 167 |
| chr17_+_43299156 | 0.09 |
ENST00000331495.3
|
FMNL1
|
formin-like 1 |
| chr17_+_7211280 | 0.08 |
ENST00000419711.2
ENST00000571955.1 ENST00000573714.1 |
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr12_-_57030115 | 0.08 |
ENST00000379441.3
ENST00000179765.5 ENST00000551812.1 |
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr19_+_58790314 | 0.08 |
ENST00000196548.5
ENST00000608843.1 |
ZNF8
ZNF8
|
Zinc finger protein 8 zinc finger protein 8 |
| chr3_+_137717571 | 0.08 |
ENST00000343735.4
|
CLDN18
|
claudin 18 |
| chrX_+_90689810 | 0.08 |
ENST00000312600.3
|
PABPC5
|
poly(A) binding protein, cytoplasmic 5 |
| chr11_+_10477733 | 0.08 |
ENST00000528723.1
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr9_-_34397800 | 0.08 |
ENST00000297623.2
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr7_-_151107106 | 0.08 |
ENST00000334493.6
|
WDR86
|
WD repeat domain 86 |
| chr1_+_209941942 | 0.08 |
ENST00000487271.1
ENST00000477431.1 |
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr19_-_30199516 | 0.08 |
ENST00000591243.1
|
C19orf12
|
chromosome 19 open reading frame 12 |
| chr3_+_141496990 | 0.08 |
ENST00000264952.2
|
GRK7
|
G protein-coupled receptor kinase 7 |
| chr14_+_29236269 | 0.08 |
ENST00000313071.4
|
FOXG1
|
forkhead box G1 |
| chr2_-_25016251 | 0.08 |
ENST00000328379.5
|
PTRHD1
|
peptidyl-tRNA hydrolase domain containing 1 |
| chr14_-_64971288 | 0.08 |
ENST00000394715.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chr17_-_76778339 | 0.08 |
ENST00000591455.1
ENST00000446868.3 ENST00000361101.4 ENST00000589296.1 |
CYTH1
|
cytohesin 1 |
| chr15_+_74528630 | 0.07 |
ENST00000398814.3
|
CCDC33
|
coiled-coil domain containing 33 |
| chrX_-_153236620 | 0.07 |
ENST00000369984.4
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr3_+_137728842 | 0.07 |
ENST00000183605.5
|
CLDN18
|
claudin 18 |
| chr7_+_149412094 | 0.07 |
ENST00000255992.10
ENST00000319551.8 |
KRBA1
|
KRAB-A domain containing 1 |
| chr5_+_161494770 | 0.07 |
ENST00000414552.2
ENST00000361925.4 |
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr11_-_46142948 | 0.07 |
ENST00000257821.4
|
PHF21A
|
PHD finger protein 21A |
| chrM_+_7586 | 0.07 |
ENST00000361739.1
|
MT-CO2
|
mitochondrially encoded cytochrome c oxidase II |
| chr22_-_30783356 | 0.07 |
ENST00000382363.3
|
RNF215
|
ring finger protein 215 |
| chr1_-_156023580 | 0.07 |
ENST00000368309.3
|
UBQLN4
|
ubiquilin 4 |
| chr12_-_117175819 | 0.07 |
ENST00000261318.3
ENST00000536380.1 |
C12orf49
|
chromosome 12 open reading frame 49 |
| chr11_+_45168182 | 0.07 |
ENST00000526442.1
|
PRDM11
|
PR domain containing 11 |
| chr12_-_45270151 | 0.07 |
ENST00000429094.2
|
NELL2
|
NEL-like 2 (chicken) |
| chr19_+_50354462 | 0.07 |
ENST00000601675.1
|
PTOV1
|
prostate tumor overexpressed 1 |
| chr19_+_1495362 | 0.07 |
ENST00000395479.4
|
REEP6
|
receptor accessory protein 6 |
| chr15_-_26108355 | 0.07 |
ENST00000356865.6
|
ATP10A
|
ATPase, class V, type 10A |
| chr12_-_31477072 | 0.07 |
ENST00000454658.2
|
FAM60A
|
family with sequence similarity 60, member A |
| chr6_+_163148973 | 0.07 |
ENST00000366888.2
|
PACRG
|
PARK2 co-regulated |
| chrX_-_23761317 | 0.07 |
ENST00000492081.1
ENST00000379303.5 ENST00000336430.7 |
ACOT9
|
acyl-CoA thioesterase 9 |
| chr6_-_31619892 | 0.07 |
ENST00000454165.1
ENST00000428326.1 ENST00000452994.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr9_+_8858102 | 0.06 |
ENST00000447950.1
ENST00000430766.1 |
RP11-75C9.1
|
RP11-75C9.1 |
| chr8_+_30244580 | 0.06 |
ENST00000523115.1
ENST00000519647.1 |
RBPMS
|
RNA binding protein with multiple splicing |
| chr20_+_2082494 | 0.06 |
ENST00000246032.3
|
STK35
|
serine/threonine kinase 35 |
| chr20_+_35202909 | 0.06 |
ENST00000558530.1
ENST00000558028.1 ENST00000560025.1 |
TGIF2-C20orf24
TGIF2
|
TGIF2-C20orf24 readthrough TGFB-induced factor homeobox 2 |
| chr7_-_150777874 | 0.06 |
ENST00000540185.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr2_-_68694390 | 0.06 |
ENST00000377957.3
|
FBXO48
|
F-box protein 48 |
| chr3_+_67048721 | 0.06 |
ENST00000295568.4
ENST00000484414.1 ENST00000460576.1 ENST00000417314.2 |
KBTBD8
|
kelch repeat and BTB (POZ) domain containing 8 |
| chr11_-_46142615 | 0.06 |
ENST00000529734.1
ENST00000323180.6 |
PHF21A
|
PHD finger protein 21A |
| chr21_-_28217721 | 0.06 |
ENST00000284984.3
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr3_+_113616317 | 0.06 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr1_+_226736446 | 0.06 |
ENST00000366788.3
ENST00000366789.4 |
C1orf95
|
chromosome 1 open reading frame 95 |
| chr9_+_131219250 | 0.06 |
ENST00000470061.1
|
ODF2
|
outer dense fiber of sperm tails 2 |
| chr19_-_6424783 | 0.06 |
ENST00000398148.3
|
KHSRP
|
KH-type splicing regulatory protein |
| chr19_-_13617247 | 0.06 |
ENST00000573710.2
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr15_-_75743915 | 0.06 |
ENST00000394949.4
|
SIN3A
|
SIN3 transcription regulator family member A |
| chr11_+_76493294 | 0.06 |
ENST00000533752.1
|
TSKU
|
tsukushi, small leucine rich proteoglycan |
| chr1_+_36690011 | 0.06 |
ENST00000354618.5
ENST00000469141.2 ENST00000478853.1 |
THRAP3
|
thyroid hormone receptor associated protein 3 |
| chr6_-_29595779 | 0.06 |
ENST00000355973.3
ENST00000377012.4 |
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr19_+_36208877 | 0.06 |
ENST00000420124.1
ENST00000222270.7 ENST00000341701.1 |
KMT2B
|
Histone-lysine N-methyltransferase 2B |
| chr1_-_156460391 | 0.05 |
ENST00000360595.3
|
MEF2D
|
myocyte enhancer factor 2D |
| chr1_-_115053781 | 0.05 |
ENST00000358465.2
ENST00000369543.2 |
TRIM33
|
tripartite motif containing 33 |
| chr11_-_64901900 | 0.05 |
ENST00000526060.1
ENST00000307289.6 ENST00000528487.1 |
SYVN1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr7_-_105516923 | 0.05 |
ENST00000478915.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr20_+_32782375 | 0.05 |
ENST00000568305.1
|
ASIP
|
agouti signaling protein |
| chr3_+_187871060 | 0.05 |
ENST00000448637.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr11_-_12030681 | 0.05 |
ENST00000529338.1
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr22_-_28197486 | 0.05 |
ENST00000302326.4
|
MN1
|
meningioma (disrupted in balanced translocation) 1 |
| chr10_+_104404644 | 0.05 |
ENST00000462202.2
|
TRIM8
|
tripartite motif containing 8 |
| chr15_-_38852251 | 0.05 |
ENST00000558432.1
|
RASGRP1
|
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
| chr12_+_120933859 | 0.05 |
ENST00000242577.6
ENST00000548214.1 ENST00000392508.2 |
DYNLL1
|
dynein, light chain, LC8-type 1 |
| chr11_-_64901978 | 0.05 |
ENST00000294256.8
ENST00000377190.3 |
SYVN1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr1_+_47264711 | 0.05 |
ENST00000371923.4
ENST00000271153.4 ENST00000371919.4 |
CYP4B1
|
cytochrome P450, family 4, subfamily B, polypeptide 1 |
| chr9_+_131218336 | 0.04 |
ENST00000372814.3
|
ODF2
|
outer dense fiber of sperm tails 2 |
| chr16_+_56899114 | 0.04 |
ENST00000566786.1
ENST00000438926.2 ENST00000563236.1 ENST00000262502.5 |
SLC12A3
|
solute carrier family 12 (sodium/chloride transporter), member 3 |
| chr17_+_38497640 | 0.04 |
ENST00000394086.3
|
RARA
|
retinoic acid receptor, alpha |
| chr2_+_25015968 | 0.04 |
ENST00000380834.2
ENST00000473706.1 |
CENPO
|
centromere protein O |
| chr16_+_28834303 | 0.04 |
ENST00000340394.8
ENST00000325215.6 ENST00000395547.2 ENST00000336783.4 ENST00000382686.4 ENST00000564304.1 |
ATXN2L
|
ataxin 2-like |
| chr22_+_38201114 | 0.04 |
ENST00000340857.2
|
H1F0
|
H1 histone family, member 0 |
| chr2_-_208030647 | 0.04 |
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr5_+_175792459 | 0.04 |
ENST00000310389.5
|
ARL10
|
ADP-ribosylation factor-like 10 |
| chr1_-_202776392 | 0.04 |
ENST00000235790.4
|
KDM5B
|
lysine (K)-specific demethylase 5B |
| chr3_+_4535025 | 0.04 |
ENST00000302640.8
ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr21_+_34804479 | 0.04 |
ENST00000421802.1
|
IFNGR2
|
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr11_-_11643540 | 0.04 |
ENST00000227756.4
|
GALNT18
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 18 |
| chr20_+_30640004 | 0.04 |
ENST00000520553.1
ENST00000518730.1 ENST00000375852.2 |
HCK
|
hemopoietic cell kinase |
| chr16_+_19183671 | 0.04 |
ENST00000562711.2
|
SYT17
|
synaptotagmin XVII |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZIC2_GLI1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.1 | 0.6 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.1 | 0.5 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.1 | 0.4 | GO:0052027 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 0.3 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 0.3 | GO:0045872 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 0.3 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) inner medullary collecting duct development(GO:0072061) |
| 0.1 | 0.2 | GO:0002586 | regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.1 | 0.3 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.1 | 0.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 0.3 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 0.1 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.1 | 0.5 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.1 | 0.4 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.1 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.0 | 0.1 | GO:2000366 | regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.0 | 0.2 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.2 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.2 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) chromatin maintenance(GO:0070827) |
| 0.0 | 0.1 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.0 | 0.2 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.1 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.0 | 0.2 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0097168 | transforming growth factor beta activation(GO:0036363) mesenchymal stem cell proliferation(GO:0097168) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.0 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.2 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.2 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.0 | 0.1 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.0 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.0 | 0.0 | GO:2000864 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0051595 | response to methylglyoxal(GO:0051595) negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.1 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.1 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.2 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.3 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 0.3 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.0 | 0.5 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.3 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.1 | 0.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.3 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.0 | 0.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.0 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.0 | 0.1 | GO:0031782 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.2 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.0 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 0.0 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.4 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |