Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
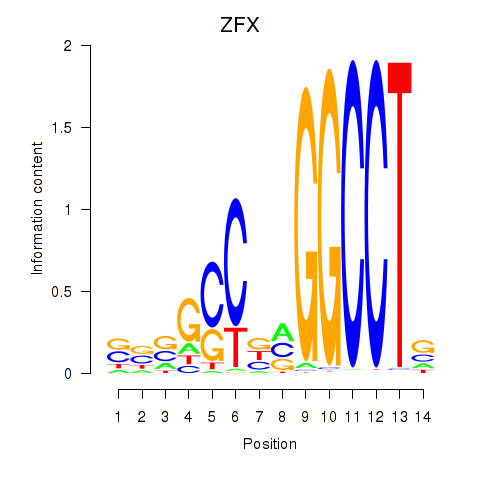
Results for ZFX
Z-value: 0.48
Transcription factors associated with ZFX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZFX
|
ENSG00000005889.11 | zinc finger protein X-linked |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZFX | hg19_v2_chrX_+_24167828_24167907 | 0.95 | 5.1e-02 | Click! |
Activity profile of ZFX motif
Sorted Z-values of ZFX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_206950095 | 0.30 |
ENST00000435627.1
|
AC007383.3
|
AC007383.3 |
| chr20_+_47662805 | 0.25 |
ENST00000262982.2
ENST00000542325.1 |
CSE1L
|
CSE1 chromosome segregation 1-like (yeast) |
| chr18_+_29672573 | 0.24 |
ENST00000578107.1
ENST00000257190.5 ENST00000580499.1 |
RNF138
|
ring finger protein 138, E3 ubiquitin protein ligase |
| chr10_+_102729249 | 0.23 |
ENST00000519649.1
ENST00000518124.1 |
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr2_-_86850949 | 0.21 |
ENST00000237455.4
|
RNF103
|
ring finger protein 103 |
| chr12_+_77158021 | 0.20 |
ENST00000550876.1
|
ZDHHC17
|
zinc finger, DHHC-type containing 17 |
| chr16_-_66952779 | 0.20 |
ENST00000570262.1
ENST00000394055.3 ENST00000299752.4 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr12_-_122241812 | 0.19 |
ENST00000538335.1
|
AC084018.1
|
AC084018.1 |
| chr10_+_22610876 | 0.19 |
ENST00000442508.1
|
BMI1
|
BMI1 polycomb ring finger oncogene |
| chr16_+_67063142 | 0.18 |
ENST00000412916.2
|
CBFB
|
core-binding factor, beta subunit |
| chr16_-_66952742 | 0.18 |
ENST00000565235.2
ENST00000568632.1 ENST00000565796.1 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr10_+_69644404 | 0.18 |
ENST00000212015.6
|
SIRT1
|
sirtuin 1 |
| chr10_+_99344071 | 0.17 |
ENST00000370647.4
ENST00000370646.4 |
HOGA1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr1_-_247171347 | 0.17 |
ENST00000339986.7
ENST00000487338.2 |
ZNF695
|
zinc finger protein 695 |
| chr9_+_33817461 | 0.16 |
ENST00000263228.3
|
UBE2R2
|
ubiquitin-conjugating enzyme E2R 2 |
| chr5_+_162864575 | 0.16 |
ENST00000512163.1
ENST00000393929.1 ENST00000340828.2 ENST00000511683.2 ENST00000510097.1 ENST00000511490.2 ENST00000510664.1 |
CCNG1
|
cyclin G1 |
| chr10_+_181418 | 0.16 |
ENST00000403354.1
ENST00000381607.4 ENST00000402736.1 |
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr8_-_103251274 | 0.16 |
ENST00000251810.3
|
RRM2B
|
ribonucleotide reductase M2 B (TP53 inducible) |
| chr1_+_26438289 | 0.15 |
ENST00000374271.4
ENST00000374269.1 |
PDIK1L
|
PDLIM1 interacting kinase 1 like |
| chr14_-_100046444 | 0.15 |
ENST00000554996.1
|
CCDC85C
|
coiled-coil domain containing 85C |
| chr4_+_53525573 | 0.15 |
ENST00000503051.1
|
USP46-AS1
|
USP46 antisense RNA 1 |
| chr5_+_78908388 | 0.15 |
ENST00000296783.3
|
PAPD4
|
PAP associated domain containing 4 |
| chr21_-_40685536 | 0.14 |
ENST00000341322.4
|
BRWD1
|
bromodomain and WD repeat domain containing 1 |
| chr2_+_223726281 | 0.14 |
ENST00000413316.1
|
ACSL3
|
acyl-CoA synthetase long-chain family member 3 |
| chr4_-_103748271 | 0.14 |
ENST00000343106.5
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr19_-_49568311 | 0.14 |
ENST00000595857.1
ENST00000451356.2 |
NTF4
|
neurotrophin 4 |
| chr1_+_28099700 | 0.14 |
ENST00000440806.2
|
STX12
|
syntaxin 12 |
| chr19_-_39390440 | 0.14 |
ENST00000249396.7
ENST00000414941.1 ENST00000392081.2 |
SIRT2
|
sirtuin 2 |
| chr10_+_119000604 | 0.14 |
ENST00000298472.5
|
SLC18A2
|
solute carrier family 18 (vesicular monoamine transporter), member 2 |
| chr13_-_111567353 | 0.14 |
ENST00000310847.4
ENST00000267339.2 ENST00000375758.5 |
ANKRD10
|
ankyrin repeat domain 10 |
| chr9_-_140115775 | 0.13 |
ENST00000391553.1
ENST00000392827.1 |
RNF208
|
ring finger protein 208 |
| chr1_-_109618566 | 0.13 |
ENST00000338366.5
|
TAF13
|
TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa |
| chr8_-_27695552 | 0.13 |
ENST00000522944.1
ENST00000301905.4 |
PBK
|
PDZ binding kinase |
| chr2_-_38604398 | 0.13 |
ENST00000443098.1
ENST00000449130.1 ENST00000378954.4 ENST00000539122.1 ENST00000419554.2 ENST00000451483.1 ENST00000406122.1 |
ATL2
|
atlastin GTPase 2 |
| chr1_+_100598691 | 0.13 |
ENST00000370143.1
ENST00000370141.2 |
TRMT13
|
tRNA methyltransferase 13 homolog (S. cerevisiae) |
| chr19_+_16435625 | 0.13 |
ENST00000248071.5
ENST00000592003.1 |
KLF2
|
Kruppel-like factor 2 |
| chr19_+_13228917 | 0.13 |
ENST00000586171.1
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr14_+_93673574 | 0.13 |
ENST00000554232.1
ENST00000556871.1 ENST00000555113.1 |
UBR7
|
ubiquitin protein ligase E3 component n-recognin 7 (putative) |
| chr10_+_88854926 | 0.13 |
ENST00000298784.1
ENST00000298786.4 |
FAM35A
|
family with sequence similarity 35, member A |
| chr15_-_49447771 | 0.12 |
ENST00000558843.1
ENST00000542928.1 ENST00000561248.1 |
COPS2
|
COP9 signalosome subunit 2 |
| chr1_+_45274154 | 0.12 |
ENST00000450269.1
ENST00000453418.1 ENST00000409335.2 |
BTBD19
|
BTB (POZ) domain containing 19 |
| chr7_-_108210048 | 0.12 |
ENST00000415914.3
ENST00000438865.1 |
THAP5
|
THAP domain containing 5 |
| chr2_-_48132924 | 0.12 |
ENST00000403359.3
|
FBXO11
|
F-box protein 11 |
| chr9_+_99212403 | 0.12 |
ENST00000375251.3
ENST00000375249.4 |
HABP4
|
hyaluronan binding protein 4 |
| chr11_-_17410629 | 0.12 |
ENST00000526912.1
|
KCNJ11
|
potassium inwardly-rectifying channel, subfamily J, member 11 |
| chr2_-_48132814 | 0.12 |
ENST00000316377.4
ENST00000378314.3 |
FBXO11
|
F-box protein 11 |
| chr22_-_41985865 | 0.12 |
ENST00000216259.7
|
PMM1
|
phosphomannomutase 1 |
| chr9_-_33264676 | 0.12 |
ENST00000472232.3
ENST00000379704.2 |
BAG1
|
BCL2-associated athanogene |
| chr20_-_32580924 | 0.12 |
ENST00000432859.1
|
RP5-1125A11.1
|
RP5-1125A11.1 |
| chr8_-_143859197 | 0.12 |
ENST00000395192.2
|
LYNX1
|
Ly6/neurotoxin 1 |
| chr3_-_37034702 | 0.12 |
ENST00000322716.5
|
EPM2AIP1
|
EPM2A (laforin) interacting protein 1 |
| chr8_-_101321584 | 0.12 |
ENST00000523167.1
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr19_-_39390350 | 0.12 |
ENST00000447739.1
ENST00000358931.5 ENST00000407552.1 |
SIRT2
|
sirtuin 2 |
| chr19_-_36523529 | 0.12 |
ENST00000593074.1
|
CLIP3
|
CAP-GLY domain containing linker protein 3 |
| chr19_+_49127600 | 0.12 |
ENST00000601704.1
ENST00000593308.1 |
SPHK2
|
sphingosine kinase 2 |
| chr10_+_134351319 | 0.12 |
ENST00000368594.3
ENST00000368593.3 |
INPP5A
|
inositol polyphosphate-5-phosphatase, 40kDa |
| chr11_-_69294647 | 0.12 |
ENST00000542064.1
|
AP000439.3
|
AP000439.3 |
| chr4_+_88928777 | 0.12 |
ENST00000237596.2
|
PKD2
|
polycystic kidney disease 2 (autosomal dominant) |
| chrX_-_71458802 | 0.11 |
ENST00000373657.1
ENST00000334463.3 |
ERCC6L
|
excision repair cross-complementing rodent repair deficiency, complementation group 6-like |
| chr16_-_66907139 | 0.11 |
ENST00000561579.2
|
NAE1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr9_-_6007787 | 0.11 |
ENST00000399933.3
ENST00000381461.2 ENST00000513355.2 |
KIAA2026
|
KIAA2026 |
| chr11_+_33037652 | 0.11 |
ENST00000311388.3
|
DEPDC7
|
DEP domain containing 7 |
| chrX_+_40440146 | 0.11 |
ENST00000535539.1
ENST00000378438.4 ENST00000436783.1 ENST00000544975.1 ENST00000535777.1 ENST00000447485.1 ENST00000423649.1 |
ATP6AP2
|
ATPase, H+ transporting, lysosomal accessory protein 2 |
| chr16_+_29817841 | 0.11 |
ENST00000322945.6
ENST00000562337.1 ENST00000566906.2 ENST00000563402.1 ENST00000219782.6 |
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr3_+_141205852 | 0.11 |
ENST00000286364.3
ENST00000452898.1 |
RASA2
|
RAS p21 protein activator 2 |
| chr6_-_138428613 | 0.11 |
ENST00000421351.3
|
PERP
|
PERP, TP53 apoptosis effector |
| chr10_+_22610124 | 0.11 |
ENST00000376663.3
|
BMI1
|
BMI1 polycomb ring finger oncogene |
| chr2_+_162101247 | 0.11 |
ENST00000439050.1
ENST00000436506.1 |
AC009299.3
|
AC009299.3 |
| chr8_-_54934708 | 0.11 |
ENST00000520534.1
ENST00000518784.1 ENST00000522635.1 |
TCEA1
|
transcription elongation factor A (SII), 1 |
| chr10_-_100027943 | 0.11 |
ENST00000260702.3
|
LOXL4
|
lysyl oxidase-like 4 |
| chrX_-_131352152 | 0.11 |
ENST00000342983.2
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr4_-_110651143 | 0.11 |
ENST00000243501.5
|
PLA2G12A
|
phospholipase A2, group XIIA |
| chr2_-_160143158 | 0.10 |
ENST00000409124.1
ENST00000358147.4 |
WDSUB1
|
WD repeat, sterile alpha motif and U-box domain containing 1 |
| chr14_-_35183755 | 0.10 |
ENST00000555765.1
|
CFL2
|
cofilin 2 (muscle) |
| chr4_+_57774042 | 0.10 |
ENST00000309042.7
|
REST
|
RE1-silencing transcription factor |
| chr16_+_67063036 | 0.10 |
ENST00000290858.6
ENST00000564034.1 |
CBFB
|
core-binding factor, beta subunit |
| chr18_+_51795774 | 0.10 |
ENST00000579534.1
ENST00000406285.3 ENST00000577612.1 ENST00000579434.1 ENST00000583136.1 |
POLI
|
polymerase (DNA directed) iota |
| chr18_+_596982 | 0.10 |
ENST00000579912.1
ENST00000400606.2 ENST00000540035.1 |
CLUL1
|
clusterin-like 1 (retinal) |
| chr9_+_129677039 | 0.10 |
ENST00000259351.5
ENST00000424082.2 ENST00000394022.3 ENST00000394011.3 ENST00000319107.4 |
RALGPS1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr19_+_17516494 | 0.10 |
ENST00000534306.1
|
CTD-2521M24.9
|
CTD-2521M24.9 |
| chr14_+_56046990 | 0.10 |
ENST00000438792.2
ENST00000395314.3 ENST00000395308.1 |
KTN1
|
kinectin 1 (kinesin receptor) |
| chr8_+_97657531 | 0.10 |
ENST00000519900.1
ENST00000517742.1 |
CPQ
|
carboxypeptidase Q |
| chr2_-_20101701 | 0.10 |
ENST00000402414.1
ENST00000333610.3 |
TTC32
|
tetratricopeptide repeat domain 32 |
| chr19_+_46000506 | 0.10 |
ENST00000396737.2
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr17_+_37894570 | 0.10 |
ENST00000394211.3
|
GRB7
|
growth factor receptor-bound protein 7 |
| chr8_-_143858590 | 0.10 |
ENST00000398906.1
ENST00000522929.1 |
LYNX1
|
Ly6/neurotoxin 1 |
| chr1_+_28099683 | 0.10 |
ENST00000373943.4
|
STX12
|
syntaxin 12 |
| chr19_-_13900972 | 0.10 |
ENST00000397557.1
|
AC008686.1
|
Uncharacterized protein |
| chr12_+_124457746 | 0.10 |
ENST00000392404.3
ENST00000538932.2 ENST00000337815.4 ENST00000540762.2 |
ZNF664
FAM101A
|
zinc finger protein 664 family with sequence similarity 101, member A |
| chr1_-_84543614 | 0.10 |
ENST00000605506.1
|
RP11-486G15.2
|
RP11-486G15.2 |
| chr8_-_41909496 | 0.10 |
ENST00000265713.2
ENST00000406337.1 ENST00000396930.3 ENST00000485568.1 ENST00000426524.1 |
KAT6A
|
K(lysine) acetyltransferase 6A |
| chr16_-_85146040 | 0.10 |
ENST00000539556.1
|
FAM92B
|
family with sequence similarity 92, member B |
| chr13_+_98628886 | 0.10 |
ENST00000490680.1
ENST00000539640.1 ENST00000403772.3 |
IPO5
|
importin 5 |
| chr15_+_23810903 | 0.10 |
ENST00000564592.1
|
MKRN3
|
makorin ring finger protein 3 |
| chr8_-_56685859 | 0.10 |
ENST00000523423.1
ENST00000523073.1 ENST00000519784.1 ENST00000434581.2 ENST00000519780.1 ENST00000521229.1 ENST00000522576.1 ENST00000523180.1 ENST00000522090.1 |
TMEM68
|
transmembrane protein 68 |
| chr14_+_53173890 | 0.10 |
ENST00000445930.2
|
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr1_-_51984908 | 0.10 |
ENST00000371730.2
|
EPS15
|
epidermal growth factor receptor pathway substrate 15 |
| chr17_+_4901199 | 0.10 |
ENST00000320785.5
ENST00000574165.1 |
KIF1C
|
kinesin family member 1C |
| chrX_+_146993534 | 0.10 |
ENST00000334557.6
ENST00000439526.2 ENST00000370475.4 |
FMR1
|
fragile X mental retardation 1 |
| chr8_+_17354587 | 0.10 |
ENST00000494857.1
ENST00000522656.1 |
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr17_-_5095126 | 0.10 |
ENST00000576772.1
ENST00000575779.1 |
ZNF594
|
zinc finger protein 594 |
| chr8_-_101322132 | 0.10 |
ENST00000523481.1
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr1_+_1260598 | 0.09 |
ENST00000488011.1
|
GLTPD1
|
glycolipid transfer protein domain containing 1 |
| chr2_-_45162783 | 0.09 |
ENST00000432125.2
|
RP11-89K21.1
|
RP11-89K21.1 |
| chr12_+_124457670 | 0.09 |
ENST00000539644.1
|
ZNF664
|
zinc finger protein 664 |
| chr1_+_215740709 | 0.09 |
ENST00000259154.4
|
KCTD3
|
potassium channel tetramerization domain containing 3 |
| chr8_-_54935001 | 0.09 |
ENST00000396401.3
ENST00000521604.2 |
TCEA1
|
transcription elongation factor A (SII), 1 |
| chr13_-_30424821 | 0.09 |
ENST00000380680.4
|
UBL3
|
ubiquitin-like 3 |
| chr17_+_48243352 | 0.09 |
ENST00000344627.6
ENST00000262018.3 ENST00000543315.1 ENST00000451235.2 ENST00000511303.1 |
SGCA
|
sarcoglycan, alpha (50kDa dystrophin-associated glycoprotein) |
| chr16_+_28874345 | 0.09 |
ENST00000566209.1
|
SH2B1
|
SH2B adaptor protein 1 |
| chr9_+_136243117 | 0.09 |
ENST00000426926.2
ENST00000371957.3 |
C9orf96
|
chromosome 9 open reading frame 96 |
| chr7_+_94536898 | 0.09 |
ENST00000433360.1
ENST00000340694.4 ENST00000424654.1 |
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr8_+_17354617 | 0.09 |
ENST00000470360.1
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr5_+_41925325 | 0.09 |
ENST00000296812.2
ENST00000281623.3 ENST00000509134.1 |
FBXO4
|
F-box protein 4 |
| chr16_+_46723552 | 0.09 |
ENST00000219097.2
ENST00000568364.2 |
ORC6
|
origin recognition complex, subunit 6 |
| chr15_-_34502197 | 0.09 |
ENST00000557877.1
|
KATNBL1
|
katanin p80 subunit B-like 1 |
| chr22_+_20850171 | 0.09 |
ENST00000445987.1
|
MED15
|
mediator complex subunit 15 |
| chr10_+_99332198 | 0.09 |
ENST00000307518.5
ENST00000298808.5 ENST00000370655.1 |
ANKRD2
|
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr3_+_27754397 | 0.09 |
ENST00000606069.1
|
RP11-222K16.2
|
RP11-222K16.2 |
| chr14_+_32546485 | 0.09 |
ENST00000345122.3
ENST00000432921.1 ENST00000433497.1 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr1_+_109289279 | 0.09 |
ENST00000370008.3
|
STXBP3
|
syntaxin binding protein 3 |
| chr5_+_76506706 | 0.09 |
ENST00000340978.3
ENST00000346042.3 ENST00000264917.5 ENST00000342343.4 ENST00000333194.4 |
PDE8B
|
phosphodiesterase 8B |
| chr9_-_123638633 | 0.09 |
ENST00000456291.1
|
PHF19
|
PHD finger protein 19 |
| chr10_+_116581503 | 0.09 |
ENST00000369248.4
ENST00000369250.3 ENST00000369246.1 |
FAM160B1
|
family with sequence similarity 160, member B1 |
| chr5_-_114880533 | 0.09 |
ENST00000274457.3
|
FEM1C
|
fem-1 homolog c (C. elegans) |
| chr15_+_91073193 | 0.09 |
ENST00000560098.1
ENST00000268184.6 |
CRTC3
|
CREB regulated transcription coactivator 3 |
| chr1_+_97187318 | 0.09 |
ENST00000609116.1
ENST00000370198.1 ENST00000370197.1 ENST00000426398.2 ENST00000394184.3 |
PTBP2
|
polypyrimidine tract binding protein 2 |
| chr3_+_19988885 | 0.09 |
ENST00000422242.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr9_+_139871948 | 0.09 |
ENST00000224167.2
ENST00000457950.1 ENST00000371625.3 ENST00000371623.3 |
PTGDS
|
prostaglandin D2 synthase 21kDa (brain) |
| chr6_-_85473073 | 0.09 |
ENST00000606621.1
|
TBX18
|
T-box 18 |
| chr2_-_241500168 | 0.09 |
ENST00000443318.1
ENST00000411765.1 |
ANKMY1
|
ankyrin repeat and MYND domain containing 1 |
| chr9_+_91003271 | 0.09 |
ENST00000375859.3
ENST00000541629.1 |
SPIN1
|
spindlin 1 |
| chr19_+_46000479 | 0.09 |
ENST00000456399.2
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr17_-_16557128 | 0.09 |
ENST00000423860.2
ENST00000311331.7 ENST00000583766.1 |
ZNF624
|
zinc finger protein 624 |
| chr1_-_156542328 | 0.09 |
ENST00000361170.2
|
IQGAP3
|
IQ motif containing GTPase activating protein 3 |
| chrX_+_154299753 | 0.09 |
ENST00000369459.2
ENST00000369462.1 ENST00000411985.1 ENST00000399042.1 |
BRCC3
|
BRCA1/BRCA2-containing complex, subunit 3 |
| chr15_+_96875657 | 0.09 |
ENST00000559679.1
ENST00000394171.2 |
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr9_-_140066495 | 0.09 |
ENST00000535352.2
|
TMEM210
|
transmembrane protein 210 |
| chr10_-_70092635 | 0.09 |
ENST00000309049.4
|
PBLD
|
phenazine biosynthesis-like protein domain containing |
| chr2_-_64881018 | 0.09 |
ENST00000313349.3
|
SERTAD2
|
SERTA domain containing 2 |
| chr15_-_49447835 | 0.09 |
ENST00000388901.5
ENST00000299259.6 |
COPS2
|
COP9 signalosome subunit 2 |
| chr19_+_30433372 | 0.08 |
ENST00000312051.6
|
URI1
|
URI1, prefoldin-like chaperone |
| chr14_+_104552016 | 0.08 |
ENST00000551177.1
ENST00000546892.2 ENST00000455920.2 |
ASPG
|
asparaginase homolog (S. cerevisiae) |
| chr10_-_70092671 | 0.08 |
ENST00000358769.2
ENST00000432941.1 ENST00000495025.2 |
PBLD
|
phenazine biosynthesis-like protein domain containing |
| chr2_-_198364581 | 0.08 |
ENST00000428204.1
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin) |
| chr16_+_67563250 | 0.08 |
ENST00000566907.1
|
FAM65A
|
family with sequence similarity 65, member A |
| chr20_+_34742650 | 0.08 |
ENST00000373945.1
ENST00000338074.2 |
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr16_-_18937480 | 0.08 |
ENST00000532700.2
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr5_-_14871866 | 0.08 |
ENST00000284268.6
|
ANKH
|
ANKH inorganic pyrophosphate transport regulator |
| chr12_-_89918522 | 0.08 |
ENST00000529983.2
|
GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) |
| chr7_+_91570240 | 0.08 |
ENST00000394564.1
|
AKAP9
|
A kinase (PRKA) anchor protein 9 |
| chr11_-_10920838 | 0.08 |
ENST00000503469.2
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr6_+_116892530 | 0.08 |
ENST00000466444.2
ENST00000368590.5 ENST00000392526.1 |
RWDD1
|
RWD domain containing 1 |
| chr14_-_23770683 | 0.08 |
ENST00000561437.1
ENST00000559942.1 ENST00000560913.1 ENST00000559314.1 ENST00000558058.1 |
PPP1R3E
|
protein phosphatase 1, regulatory subunit 3E |
| chr1_+_18807424 | 0.08 |
ENST00000400664.1
|
KLHDC7A
|
kelch domain containing 7A |
| chr8_-_103250997 | 0.08 |
ENST00000522368.1
|
RRM2B
|
ribonucleotide reductase M2 B (TP53 inducible) |
| chr19_-_6481776 | 0.08 |
ENST00000543576.1
ENST00000590173.1 ENST00000381480.2 |
DENND1C
|
DENN/MADD domain containing 1C |
| chr9_-_6015607 | 0.08 |
ENST00000259569.5
|
RANBP6
|
RAN binding protein 6 |
| chr19_-_53466095 | 0.08 |
ENST00000391786.2
ENST00000434371.2 ENST00000357666.4 ENST00000438970.2 ENST00000270457.4 ENST00000535506.1 ENST00000444460.2 ENST00000457013.2 |
ZNF816
|
zinc finger protein 816 |
| chr1_-_54519134 | 0.08 |
ENST00000371341.1
|
TMEM59
|
transmembrane protein 59 |
| chr8_-_145691031 | 0.08 |
ENST00000424149.2
ENST00000530637.1 ENST00000306145.5 |
CYHR1
|
cysteine/histidine-rich 1 |
| chr19_+_38893809 | 0.08 |
ENST00000589408.1
|
FAM98C
|
family with sequence similarity 98, member C |
| chr1_-_51984990 | 0.08 |
ENST00000371733.3
|
EPS15
|
epidermal growth factor receptor pathway substrate 15 |
| chr19_-_36297348 | 0.08 |
ENST00000589835.1
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr7_-_32534850 | 0.08 |
ENST00000409952.3
ENST00000409909.3 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr6_+_31126291 | 0.08 |
ENST00000376257.3
ENST00000376255.4 |
TCF19
|
transcription factor 19 |
| chr2_+_48010221 | 0.08 |
ENST00000234420.5
|
MSH6
|
mutS homolog 6 |
| chr5_+_78908233 | 0.08 |
ENST00000453514.1
ENST00000423041.2 ENST00000504233.1 ENST00000428308.2 |
PAPD4
|
PAP associated domain containing 4 |
| chr15_-_59225758 | 0.08 |
ENST00000558486.1
ENST00000560682.1 ENST00000249736.7 ENST00000559880.1 ENST00000536328.1 |
SLTM
|
SAFB-like, transcription modulator |
| chr19_+_36705504 | 0.08 |
ENST00000456324.1
|
ZNF146
|
zinc finger protein 146 |
| chr20_-_50159198 | 0.08 |
ENST00000371564.3
ENST00000396009.3 ENST00000610033.1 |
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 |
| chr20_-_1373682 | 0.08 |
ENST00000381724.3
|
FKBP1A
|
FK506 binding protein 1A, 12kDa |
| chr9_-_33264557 | 0.08 |
ENST00000473781.1
ENST00000488499.1 |
BAG1
|
BCL2-associated athanogene |
| chr7_+_99156314 | 0.08 |
ENST00000425063.1
ENST00000493277.1 |
ZNF655
|
zinc finger protein 655 |
| chr12_+_8185288 | 0.08 |
ENST00000162391.3
|
FOXJ2
|
forkhead box J2 |
| chr4_-_174255536 | 0.08 |
ENST00000446922.2
|
HMGB2
|
high mobility group box 2 |
| chr12_-_121973974 | 0.08 |
ENST00000538379.1
ENST00000541318.1 ENST00000541511.1 |
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr15_+_40763150 | 0.08 |
ENST00000306243.5
ENST00000559991.1 |
CHST14
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 14 |
| chr6_+_116892641 | 0.08 |
ENST00000487832.2
ENST00000518117.1 |
RWDD1
|
RWD domain containing 1 |
| chr2_-_160568918 | 0.08 |
ENST00000453016.1
ENST00000607836.1 |
AC009961.3
|
AC009961.3 |
| chr5_-_693500 | 0.08 |
ENST00000360578.5
|
TPPP
|
tubulin polymerization promoting protein |
| chr14_+_64971292 | 0.08 |
ENST00000358738.3
ENST00000394712.2 |
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr4_-_110651111 | 0.08 |
ENST00000502283.1
|
PLA2G12A
|
phospholipase A2, group XIIA |
| chr19_+_3359561 | 0.08 |
ENST00000589123.1
ENST00000346156.5 ENST00000395111.3 ENST00000586919.1 |
NFIC
|
nuclear factor I/C (CCAAT-binding transcription factor) |
| chr14_+_56046914 | 0.08 |
ENST00000413890.2
ENST00000395309.3 ENST00000554567.1 ENST00000555498.1 |
KTN1
|
kinectin 1 (kinesin receptor) |
| chr11_-_68609377 | 0.08 |
ENST00000265641.5
ENST00000376618.2 |
CPT1A
|
carnitine palmitoyltransferase 1A (liver) |
| chr16_+_47495225 | 0.08 |
ENST00000299167.8
ENST00000323584.5 ENST00000563376.1 |
PHKB
|
phosphorylase kinase, beta |
| chr9_-_95527079 | 0.08 |
ENST00000356884.6
ENST00000375512.3 |
BICD2
|
bicaudal D homolog 2 (Drosophila) |
| chr15_+_40453204 | 0.08 |
ENST00000287598.6
ENST00000412359.3 |
BUB1B
|
BUB1 mitotic checkpoint serine/threonine kinase B |
| chr1_-_226496772 | 0.08 |
ENST00000359525.2
ENST00000460719.1 |
LIN9
|
lin-9 homolog (C. elegans) |
| chr16_+_67562702 | 0.08 |
ENST00000379312.3
ENST00000042381.4 ENST00000540839.3 |
FAM65A
|
family with sequence similarity 65, member A |
| chr1_-_59165763 | 0.07 |
ENST00000472487.1
|
MYSM1
|
Myb-like, SWIRM and MPN domains 1 |
| chr14_-_75422280 | 0.07 |
ENST00000238607.6
ENST00000553716.1 |
PGF
|
placental growth factor |
| chr17_+_37618257 | 0.07 |
ENST00000447079.4
|
CDK12
|
cyclin-dependent kinase 12 |
| chr17_-_15902903 | 0.07 |
ENST00000486655.1
|
ZSWIM7
|
zinc finger, SWIM-type containing 7 |
| chr2_+_85646054 | 0.07 |
ENST00000389938.2
|
SH2D6
|
SH2 domain containing 6 |
| chr3_+_44903361 | 0.07 |
ENST00000302392.4
|
TMEM42
|
transmembrane protein 42 |
| chr12_-_57081940 | 0.07 |
ENST00000436399.2
|
PTGES3
|
prostaglandin E synthase 3 (cytosolic) |
| chr4_+_110354928 | 0.07 |
ENST00000504968.2
ENST00000399100.2 ENST00000265175.5 |
SEC24B
|
SEC24 family member B |
| chr9_+_129987488 | 0.07 |
ENST00000446764.1
|
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZFX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0061433 | cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 0.1 | 0.2 | GO:0010160 | sensory organ boundary specification(GO:0008052) formation of organ boundary(GO:0010160) taste bud development(GO:0061193) |
| 0.1 | 0.2 | GO:0061461 | lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine import into cell(GO:1903410) |
| 0.1 | 0.2 | GO:1990619 | histone H3-K9 deacetylation(GO:1990619) |
| 0.0 | 0.1 | GO:0045041 | B cell cytokine production(GO:0002368) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.2 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.2 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.1 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.0 | 0.2 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.0 | 0.1 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.0 | 0.1 | GO:0035261 | external genitalia morphogenesis(GO:0035261) |
| 0.0 | 0.1 | GO:2000705 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.1 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.1 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.2 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.0 | 0.1 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.0 | 0.1 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 0.1 | GO:1901254 | modulation by host of viral RNA genome replication(GO:0044830) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.0 | 0.1 | GO:2000174 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.3 | GO:0071484 | cellular response to light intensity(GO:0071484) |
| 0.0 | 0.1 | GO:0006562 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.2 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.0 | 0.1 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.1 | GO:0050960 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.0 | 0.0 | GO:0061740 | protein targeting to lysosome involved in chaperone-mediated autophagy(GO:0061740) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.1 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.1 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.0 | 0.2 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.3 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.0 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.2 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.1 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.1 | GO:0035508 | positive regulation of myosin-light-chain-phosphatase activity(GO:0035508) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.0 | 0.1 | GO:1903181 | negative regulation of late endosome to lysosome transport(GO:1902823) regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.0 | 0.2 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.1 | GO:0071314 | cellular response to cocaine(GO:0071314) cellular response to luteinizing hormone stimulus(GO:0071373) |
| 0.0 | 0.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.0 | 0.1 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.3 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.0 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.1 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.0 | 0.1 | GO:0046707 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.0 | 0.1 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) negative regulation of double-strand break repair via single-strand annealing(GO:1901291) |
| 0.0 | 0.2 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.2 | GO:0021903 | rostrocaudal neural tube patterning(GO:0021903) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.0 | GO:0042704 | uterine wall breakdown(GO:0042704) |
| 0.0 | 0.0 | GO:2000374 | regulation of oxygen metabolic process(GO:2000374) |
| 0.0 | 0.0 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.0 | 0.2 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.0 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.0 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.1 | GO:1905075 | occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.0 | 0.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.1 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.0 | 0.0 | GO:1903336 | intralumenal vesicle formation(GO:0070676) negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.1 | GO:0010748 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.3 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.1 | GO:0015960 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.1 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0036100 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.0 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.0 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.0 | GO:0000921 | septin ring assembly(GO:0000921) septin ring organization(GO:0031106) |
| 0.0 | 0.0 | GO:0002339 | B cell selection(GO:0002339) B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) apoptotic process involved in embryonic digit morphogenesis(GO:1902263) positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.0 | 0.0 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.0 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.1 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.0 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.0 | 0.0 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.0 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.0 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.2 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.1 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.1 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.2 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.0 | 0.4 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.0 | GO:0031116 | positive regulation of microtubule polymerization or depolymerization(GO:0031112) regulation of microtubule polymerization(GO:0031113) positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.0 | GO:0050758 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.1 | GO:0034184 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.0 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.0 | 0.1 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.0 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.0 | 0.0 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 0.0 | 0.0 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:0060828 | regulation of canonical Wnt signaling pathway(GO:0060828) |
| 0.0 | 0.2 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.0 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 0.1 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.0 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.1 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.1 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.0 | 0.3 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.0 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.0 | 0.4 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:1902737 | dendritic filopodium(GO:1902737) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.0 | 0.1 | GO:0000229 | cytoplasmic chromosome(GO:0000229) |
| 0.0 | 0.2 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.1 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.0 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.0 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.0 | GO:0070993 | translation preinitiation complex(GO:0070993) |
| 0.0 | 0.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.0 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.0 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.0 | 0.1 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.0 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.0 | 0.0 | GO:0032449 | CBM complex(GO:0032449) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0046970 | NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.1 | 0.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.3 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.1 | 0.4 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.1 | 0.2 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.0 | 0.3 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.2 | GO:0005292 | high-affinity basic amino acid transmembrane transporter activity(GO:0005287) high-affinity arginine transmembrane transporter activity(GO:0005289) high-affinity lysine transmembrane transporter activity(GO:0005292) |
| 0.0 | 0.1 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.0 | 0.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.2 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.1 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.1 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.1 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.1 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 0.1 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.1 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.2 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.2 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.1 | GO:0034039 | 8-oxo-7,8-dihydroguanine DNA N-glycosylase activity(GO:0034039) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.1 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.0 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.0 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.0 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.0 | 0.0 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) leukemia inhibitory factor receptor activity(GO:0004923) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.0 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.0 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 0.1 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.0 | 0.1 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.0 | GO:0047322 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
| 0.0 | 0.0 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.0 | 0.0 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.0 | 0.1 | GO:0019144 | ADP-sugar diphosphatase activity(GO:0019144) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.0 | 0.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.0 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |