Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
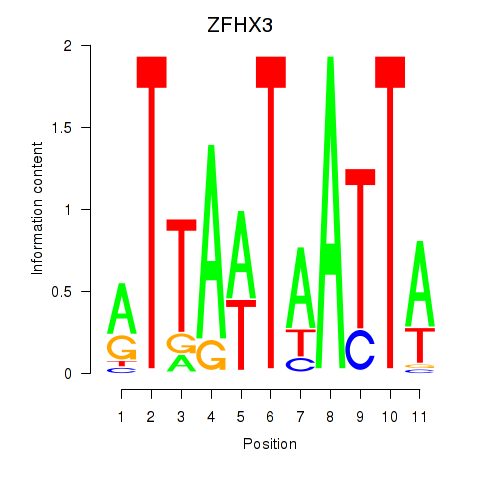
Results for ZFHX3
Z-value: 0.48
Transcription factors associated with ZFHX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZFHX3
|
ENSG00000140836.10 | zinc finger homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZFHX3 | hg19_v2_chr16_-_73082274_73082274 | -0.49 | 5.1e-01 | Click! |
Activity profile of ZFHX3 motif
Sorted Z-values of ZFHX3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_111649015 | 0.21 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr16_-_28192360 | 0.19 |
ENST00000570033.1
|
XPO6
|
exportin 6 |
| chr19_-_36304201 | 0.19 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr17_-_64225508 | 0.16 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chrM_+_14741 | 0.15 |
ENST00000361789.2
|
MT-CYB
|
mitochondrially encoded cytochrome b |
| chr12_-_91546926 | 0.14 |
ENST00000550758.1
|
DCN
|
decorin |
| chr6_+_161123270 | 0.13 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chrM_+_10053 | 0.13 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr9_+_44868935 | 0.13 |
ENST00000448436.2
|
RP11-160N1.10
|
RP11-160N1.10 |
| chr11_-_125366089 | 0.12 |
ENST00000366139.3
ENST00000278919.3 |
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr16_-_69385681 | 0.11 |
ENST00000288025.3
|
TMED6
|
transmembrane emp24 protein transport domain containing 6 |
| chr14_+_95027772 | 0.11 |
ENST00000555095.1
ENST00000298841.5 ENST00000554220.1 ENST00000553780.1 |
SERPINA4
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 4 serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr16_-_75284758 | 0.10 |
ENST00000561970.1
|
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr11_-_61687739 | 0.10 |
ENST00000531922.1
ENST00000301773.5 |
RAB3IL1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr14_-_83262540 | 0.09 |
ENST00000554451.1
|
RP11-11K13.1
|
RP11-11K13.1 |
| chr5_+_60933634 | 0.09 |
ENST00000505642.1
|
C5orf64
|
chromosome 5 open reading frame 64 |
| chr3_-_161089289 | 0.09 |
ENST00000497137.1
|
SPTSSB
|
serine palmitoyltransferase, small subunit B |
| chr16_+_87636474 | 0.09 |
ENST00000284262.2
|
JPH3
|
junctophilin 3 |
| chr1_+_76540386 | 0.08 |
ENST00000328299.3
|
ST6GALNAC3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr15_-_42343388 | 0.08 |
ENST00000399518.3
|
PLA2G4E
|
phospholipase A2, group IVE |
| chr16_-_15474904 | 0.08 |
ENST00000534094.1
|
NPIPA5
|
nuclear pore complex interacting protein family, member A5 |
| chrY_-_20935572 | 0.08 |
ENST00000382852.1
ENST00000344884.4 ENST00000304790.3 |
HSFY2
|
heat shock transcription factor, Y linked 2 |
| chrX_-_18690210 | 0.08 |
ENST00000379984.3
|
RS1
|
retinoschisin 1 |
| chr12_+_54410664 | 0.08 |
ENST00000303406.4
|
HOXC4
|
homeobox C4 |
| chrY_+_20708557 | 0.08 |
ENST00000307393.2
ENST00000309834.4 ENST00000382856.2 |
HSFY1
|
heat shock transcription factor, Y-linked 1 |
| chrX_+_1710484 | 0.07 |
ENST00000313871.3
ENST00000381261.3 |
AKAP17A
|
A kinase (PRKA) anchor protein 17A |
| chr11_+_71934962 | 0.07 |
ENST00000543234.1
|
INPPL1
|
inositol polyphosphate phosphatase-like 1 |
| chrM_+_8527 | 0.07 |
ENST00000361899.2
|
MT-ATP6
|
mitochondrially encoded ATP synthase 6 |
| chr4_-_155511887 | 0.07 |
ENST00000302053.3
ENST00000403106.3 |
FGA
|
fibrinogen alpha chain |
| chr7_-_86595190 | 0.07 |
ENST00000398276.2
ENST00000416314.1 ENST00000425689.1 |
KIAA1324L
|
KIAA1324-like |
| chr13_-_30881134 | 0.07 |
ENST00000380617.3
ENST00000441394.1 |
KATNAL1
|
katanin p60 subunit A-like 1 |
| chrM_-_14670 | 0.07 |
ENST00000361681.2
|
MT-ND6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chrX_-_110655306 | 0.06 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr19_-_58864848 | 0.06 |
ENST00000263100.3
|
A1BG
|
alpha-1-B glycoprotein |
| chr15_-_76352069 | 0.06 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chr9_-_95298314 | 0.06 |
ENST00000344604.5
ENST00000375540.1 |
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr6_+_126070726 | 0.06 |
ENST00000368364.3
|
HEY2
|
hes-related family bHLH transcription factor with YRPW motif 2 |
| chr4_-_149365827 | 0.06 |
ENST00000344721.4
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr21_+_40824003 | 0.06 |
ENST00000452550.1
|
SH3BGR
|
SH3 domain binding glutamic acid-rich protein |
| chr22_-_30970560 | 0.06 |
ENST00000402369.1
ENST00000406361.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr5_-_125930929 | 0.05 |
ENST00000553117.1
ENST00000447989.2 ENST00000409134.3 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chrM_+_9207 | 0.05 |
ENST00000362079.2
|
MT-CO3
|
mitochondrially encoded cytochrome c oxidase III |
| chr14_-_54425475 | 0.05 |
ENST00000559642.1
|
BMP4
|
bone morphogenetic protein 4 |
| chr11_+_22696314 | 0.05 |
ENST00000532398.1
ENST00000433790.1 |
GAS2
|
growth arrest-specific 2 |
| chr1_-_247921982 | 0.05 |
ENST00000408896.2
|
OR1C1
|
olfactory receptor, family 1, subfamily C, member 1 |
| chr12_+_65996599 | 0.04 |
ENST00000539116.1
ENST00000541391.1 |
RP11-221N13.3
|
RP11-221N13.3 |
| chr11_-_111649074 | 0.04 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr13_-_30880979 | 0.04 |
ENST00000414289.1
|
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr22_-_22337204 | 0.04 |
ENST00000430142.1
ENST00000357179.5 |
TOP3B
|
topoisomerase (DNA) III beta |
| chr12_+_131438443 | 0.04 |
ENST00000261654.5
|
GPR133
|
G protein-coupled receptor 133 |
| chrX_+_84258832 | 0.04 |
ENST00000373173.2
|
APOOL
|
apolipoprotein O-like |
| chr6_-_161085291 | 0.03 |
ENST00000316300.5
|
LPA
|
lipoprotein, Lp(a) |
| chr2_+_190541153 | 0.03 |
ENST00000313581.4
ENST00000438402.2 ENST00000431575.2 ENST00000281412.6 |
ANKAR
|
ankyrin and armadillo repeat containing |
| chr19_+_926000 | 0.03 |
ENST00000263620.3
|
ARID3A
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr4_+_3443614 | 0.03 |
ENST00000382774.3
ENST00000511533.1 |
HGFAC
|
HGF activator |
| chr3_+_46919235 | 0.03 |
ENST00000449590.1
|
PTH1R
|
parathyroid hormone 1 receptor |
| chr7_+_150725510 | 0.03 |
ENST00000461373.1
ENST00000358849.4 ENST00000297504.6 ENST00000542328.1 ENST00000498578.1 ENST00000356058.4 ENST00000477719.1 ENST00000477092.1 |
ABCB8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8 |
| chr2_+_105050794 | 0.03 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr17_-_26697304 | 0.03 |
ENST00000536498.1
|
VTN
|
vitronectin |
| chr2_-_60780702 | 0.03 |
ENST00000359629.5
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chr6_-_116447283 | 0.03 |
ENST00000452729.1
ENST00000243222.4 |
COL10A1
|
collagen, type X, alpha 1 |
| chr2_-_60780546 | 0.03 |
ENST00000358510.4
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chr2_-_60780536 | 0.03 |
ENST00000538214.1
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chr19_-_46234119 | 0.03 |
ENST00000317683.3
|
FBXO46
|
F-box protein 46 |
| chr1_+_63063152 | 0.02 |
ENST00000371129.3
|
ANGPTL3
|
angiopoietin-like 3 |
| chr6_-_31681839 | 0.02 |
ENST00000409239.1
ENST00000461287.1 |
LY6G6E
XXbac-BPG32J3.20
|
lymphocyte antigen 6 complex, locus G6E (pseudogene) Uncharacterized protein |
| chr1_-_159684371 | 0.02 |
ENST00000255030.5
ENST00000437342.1 ENST00000368112.1 ENST00000368111.1 ENST00000368110.1 ENST00000343919.2 |
CRP
|
C-reactive protein, pentraxin-related |
| chr3_-_49066811 | 0.02 |
ENST00000442157.1
ENST00000326739.4 |
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr10_+_45406627 | 0.02 |
ENST00000389583.4
|
TMEM72
|
transmembrane protein 72 |
| chr6_-_152793728 | 0.02 |
ENST00000495090.2
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr9_-_123812542 | 0.02 |
ENST00000223642.1
|
C5
|
complement component 5 |
| chr12_+_53835383 | 0.02 |
ENST00000429243.2
|
PRR13
|
proline rich 13 |
| chr8_+_98900132 | 0.02 |
ENST00000520016.1
|
MATN2
|
matrilin 2 |
| chr7_+_100026406 | 0.02 |
ENST00000414441.1
|
MEPCE
|
methylphosphate capping enzyme |
| chr20_-_33735070 | 0.02 |
ENST00000374491.3
ENST00000542871.1 ENST00000374492.3 |
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr2_-_60780607 | 0.02 |
ENST00000537768.1
ENST00000335712.6 ENST00000356842.4 |
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chr8_+_104831440 | 0.02 |
ENST00000515551.1
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr7_+_48211048 | 0.02 |
ENST00000435803.1
|
ABCA13
|
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr11_-_125366018 | 0.02 |
ENST00000527534.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr12_-_10321754 | 0.02 |
ENST00000539518.1
|
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr8_-_70745575 | 0.02 |
ENST00000524945.1
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1 |
| chr12_+_53835539 | 0.02 |
ENST00000547368.1
ENST00000379786.4 ENST00000551945.1 |
PRR13
|
proline rich 13 |
| chr12_+_53835508 | 0.02 |
ENST00000551003.1
ENST00000549068.1 ENST00000549740.1 ENST00000546581.1 ENST00000549581.1 ENST00000541275.1 |
PRR13
PCBP2
|
proline rich 13 poly(rC) binding protein 2 |
| chr12_+_59989791 | 0.01 |
ENST00000552432.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr19_+_50016610 | 0.01 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr7_-_7575477 | 0.01 |
ENST00000399429.3
|
COL28A1
|
collagen, type XXVIII, alpha 1 |
| chr7_-_91794590 | 0.01 |
ENST00000430130.2
ENST00000454089.2 |
LRRD1
|
leucine-rich repeats and death domain containing 1 |
| chrX_-_32173579 | 0.01 |
ENST00000359836.1
ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD
|
dystrophin |
| chr2_+_234627424 | 0.01 |
ENST00000373409.3
|
UGT1A4
|
UDP glucuronosyltransferase 1 family, polypeptide A4 |
| chrX_-_15619076 | 0.01 |
ENST00000252519.3
|
ACE2
|
angiotensin I converting enzyme 2 |
| chrX_+_9431324 | 0.01 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr15_+_58724184 | 0.01 |
ENST00000433326.2
|
LIPC
|
lipase, hepatic |
| chr2_-_87248975 | 0.01 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr6_+_25754927 | 0.01 |
ENST00000377905.4
ENST00000439485.2 |
SLC17A4
|
solute carrier family 17, member 4 |
| chr14_+_39703084 | 0.00 |
ENST00000553728.1
|
RP11-407N17.3
|
cTAGE family member 5 isoform 4 |
| chr3_-_114477787 | 0.00 |
ENST00000464560.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr8_-_18541603 | 0.00 |
ENST00000428502.2
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZFHX3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.2 | GO:0010133 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:2000173 | negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.0 | 0.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.0 | 0.1 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.1 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.2 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.1 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.1 | GO:0036025 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.1 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 0.2 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |