Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
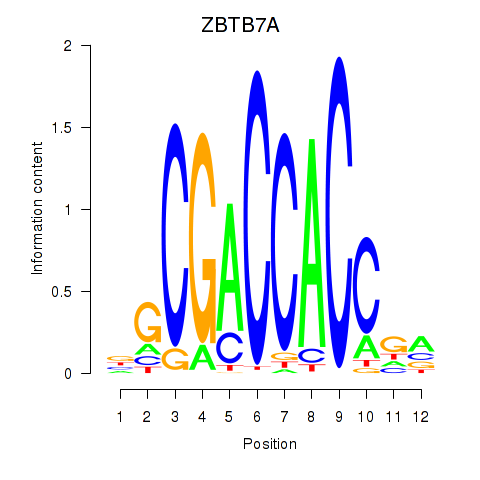
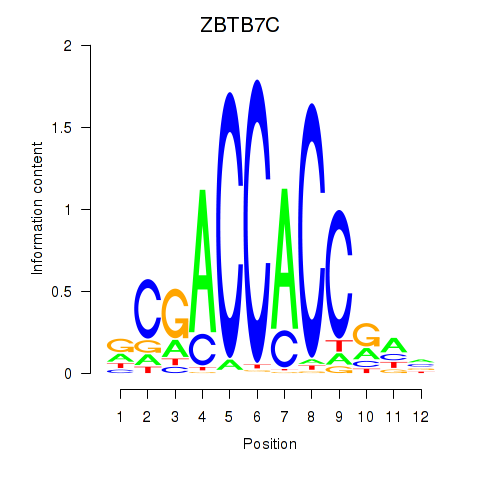
Results for ZBTB7A_ZBTB7C
Z-value: 0.39
Transcription factors associated with ZBTB7A_ZBTB7C
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB7A
|
ENSG00000178951.4 | zinc finger and BTB domain containing 7A |
|
ZBTB7C
|
ENSG00000184828.5 | zinc finger and BTB domain containing 7C |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB7A | hg19_v2_chr19_-_4065730_4065909 | -0.91 | 8.9e-02 | Click! |
| ZBTB7C | hg19_v2_chr18_-_45663666_45663732 | -0.57 | 4.3e-01 | Click! |
Activity profile of ZBTB7A_ZBTB7C motif
Sorted Z-values of ZBTB7A_ZBTB7C motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_129872672 | 0.30 |
ENST00000531431.1
ENST00000527581.1 |
PRDM10
|
PR domain containing 10 |
| chr2_+_27505260 | 0.21 |
ENST00000380075.2
ENST00000296098.4 |
TRIM54
|
tripartite motif containing 54 |
| chr19_-_39832563 | 0.18 |
ENST00000599274.1
|
CTC-246B18.10
|
CTC-246B18.10 |
| chr5_+_139493665 | 0.18 |
ENST00000331327.3
|
PURA
|
purine-rich element binding protein A |
| chr11_+_66115304 | 0.18 |
ENST00000531602.1
|
RP11-867G23.8
|
Uncharacterized protein |
| chr21_+_38338737 | 0.17 |
ENST00000430068.1
|
AP000704.5
|
AP000704.5 |
| chr15_-_64126084 | 0.17 |
ENST00000560316.1
ENST00000443617.2 ENST00000560462.1 ENST00000558532.1 ENST00000561400.1 |
HERC1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr17_+_79990058 | 0.16 |
ENST00000584341.1
|
RAC3
|
ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) |
| chr5_+_149109825 | 0.14 |
ENST00000360453.4
ENST00000394320.3 ENST00000309241.5 |
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr16_+_53164956 | 0.14 |
ENST00000563410.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr19_+_4402659 | 0.14 |
ENST00000301280.5
ENST00000585854.1 |
CHAF1A
|
chromatin assembly factor 1, subunit A (p150) |
| chr17_-_39769005 | 0.13 |
ENST00000301653.4
ENST00000593067.1 |
KRT16
|
keratin 16 |
| chr13_-_30880979 | 0.13 |
ENST00000414289.1
|
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr16_+_84002234 | 0.12 |
ENST00000305202.4
|
NECAB2
|
N-terminal EF-hand calcium binding protein 2 |
| chr4_-_170192000 | 0.12 |
ENST00000502315.1
|
SH3RF1
|
SH3 domain containing ring finger 1 |
| chr2_+_25016282 | 0.12 |
ENST00000260662.1
|
CENPO
|
centromere protein O |
| chr7_-_127672146 | 0.11 |
ENST00000476782.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr5_-_138739739 | 0.11 |
ENST00000514983.1
ENST00000507779.2 ENST00000451821.2 ENST00000450845.2 ENST00000509959.1 ENST00000302091.5 |
SPATA24
|
spermatogenesis associated 24 |
| chr14_+_100705322 | 0.11 |
ENST00000262238.4
|
YY1
|
YY1 transcription factor |
| chr13_-_30881134 | 0.11 |
ENST00000380617.3
ENST00000441394.1 |
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr14_+_29236269 | 0.11 |
ENST00000313071.4
|
FOXG1
|
forkhead box G1 |
| chr17_+_4736812 | 0.10 |
ENST00000453408.3
|
MINK1
|
misshapen-like kinase 1 |
| chr3_+_15469058 | 0.10 |
ENST00000432764.2
|
EAF1
|
ELL associated factor 1 |
| chr17_+_4736627 | 0.10 |
ENST00000355280.6
ENST00000347992.7 |
MINK1
|
misshapen-like kinase 1 |
| chr17_-_7137582 | 0.09 |
ENST00000575756.1
ENST00000575458.1 |
DVL2
|
dishevelled segment polarity protein 2 |
| chr7_-_127671674 | 0.09 |
ENST00000478726.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr17_+_73201754 | 0.09 |
ENST00000583569.1
ENST00000245544.4 ENST00000579324.1 ENST00000541827.1 ENST00000579298.1 ENST00000447371.2 |
NUP85
|
nucleoporin 85kDa |
| chr17_+_79989937 | 0.09 |
ENST00000580965.1
|
RAC3
|
ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) |
| chr1_-_156722195 | 0.08 |
ENST00000368206.5
|
HDGF
|
hepatoma-derived growth factor |
| chr3_-_71803917 | 0.08 |
ENST00000421769.2
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr17_+_40834580 | 0.08 |
ENST00000264638.4
|
CNTNAP1
|
contactin associated protein 1 |
| chr14_+_39644425 | 0.08 |
ENST00000556530.1
|
PNN
|
pinin, desmosome associated protein |
| chr11_-_17035943 | 0.08 |
ENST00000355661.3
ENST00000532079.1 ENST00000448080.2 ENST00000531066.1 |
PLEKHA7
|
pleckstrin homology domain containing, family A member 7 |
| chr9_+_137533615 | 0.08 |
ENST00000371817.3
|
COL5A1
|
collagen, type V, alpha 1 |
| chr2_-_97405775 | 0.08 |
ENST00000264963.4
ENST00000537039.1 ENST00000377079.4 ENST00000426463.2 ENST00000534882.1 |
LMAN2L
|
lectin, mannose-binding 2-like |
| chr5_-_132113063 | 0.08 |
ENST00000378719.2
|
SEPT8
|
septin 8 |
| chr10_-_101380121 | 0.08 |
ENST00000370495.4
|
SLC25A28
|
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr11_+_66045634 | 0.08 |
ENST00000528852.1
ENST00000311445.6 |
CNIH2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr17_-_56065540 | 0.07 |
ENST00000583932.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr17_-_1531635 | 0.07 |
ENST00000571650.1
|
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr1_+_53098862 | 0.07 |
ENST00000517870.1
|
FAM159A
|
family with sequence similarity 159, member A |
| chr11_-_57103327 | 0.07 |
ENST00000529002.1
ENST00000278412.2 |
SSRP1
|
structure specific recognition protein 1 |
| chr20_+_61340179 | 0.07 |
ENST00000370501.3
|
NTSR1
|
neurotensin receptor 1 (high affinity) |
| chr1_-_92952433 | 0.07 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr17_+_64961026 | 0.07 |
ENST00000262138.3
|
CACNG4
|
calcium channel, voltage-dependent, gamma subunit 4 |
| chr14_+_70346125 | 0.07 |
ENST00000361956.3
ENST00000381280.4 |
SMOC1
|
SPARC related modular calcium binding 1 |
| chr11_+_65837907 | 0.07 |
ENST00000320580.4
|
PACS1
|
phosphofurin acidic cluster sorting protein 1 |
| chr6_-_36953833 | 0.07 |
ENST00000538808.1
ENST00000460219.1 ENST00000373616.5 ENST00000373627.5 |
MTCH1
|
mitochondrial carrier 1 |
| chr12_+_57943781 | 0.07 |
ENST00000455537.2
ENST00000286452.5 |
KIF5A
|
kinesin family member 5A |
| chr17_-_42298201 | 0.06 |
ENST00000527034.1
|
UBTF
|
upstream binding transcription factor, RNA polymerase I |
| chr14_-_102771462 | 0.06 |
ENST00000522874.1
|
MOK
|
MOK protein kinase |
| chr14_+_33408449 | 0.06 |
ENST00000346562.2
ENST00000341321.4 ENST00000548645.1 ENST00000356141.4 ENST00000357798.5 |
NPAS3
|
neuronal PAS domain protein 3 |
| chr1_-_22469459 | 0.06 |
ENST00000290167.6
|
WNT4
|
wingless-type MMTV integration site family, member 4 |
| chr17_+_43209967 | 0.06 |
ENST00000431281.1
ENST00000591859.1 |
ACBD4
|
acyl-CoA binding domain containing 4 |
| chr1_+_182992545 | 0.06 |
ENST00000258341.4
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2) |
| chr4_-_16085314 | 0.06 |
ENST00000510224.1
|
PROM1
|
prominin 1 |
| chr1_-_47697387 | 0.06 |
ENST00000371884.2
|
TAL1
|
T-cell acute lymphocytic leukemia 1 |
| chr2_+_149402553 | 0.06 |
ENST00000258484.6
ENST00000409654.1 |
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr6_-_33290580 | 0.06 |
ENST00000446511.1
ENST00000446403.1 ENST00000414083.2 ENST00000266000.6 ENST00000374542.5 |
DAXX
|
death-domain associated protein |
| chr17_-_7137857 | 0.06 |
ENST00000005340.5
|
DVL2
|
dishevelled segment polarity protein 2 |
| chrX_-_51812268 | 0.06 |
ENST00000486010.1
ENST00000497164.1 ENST00000360134.6 ENST00000485287.1 ENST00000335504.5 ENST00000431659.1 |
MAGED4B
|
melanoma antigen family D, 4B |
| chr19_-_48673552 | 0.06 |
ENST00000536218.1
ENST00000596549.1 |
LIG1
|
ligase I, DNA, ATP-dependent |
| chr19_-_13044494 | 0.05 |
ENST00000593021.1
ENST00000587981.1 ENST00000423140.2 ENST00000314606.4 |
FARSA
|
phenylalanyl-tRNA synthetase, alpha subunit |
| chr1_+_110754094 | 0.05 |
ENST00000369787.3
ENST00000413138.3 ENST00000438661.2 |
KCNC4
|
potassium voltage-gated channel, Shaw-related subfamily, member 4 |
| chr12_+_132312931 | 0.05 |
ENST00000360564.1
ENST00000545671.1 ENST00000545790.1 |
MMP17
|
matrix metallopeptidase 17 (membrane-inserted) |
| chr5_-_176924562 | 0.05 |
ENST00000359895.2
ENST00000355572.2 ENST00000355841.2 ENST00000393551.1 ENST00000505074.1 ENST00000356618.4 ENST00000393546.4 |
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
| chr19_-_59070239 | 0.05 |
ENST00000595957.1
ENST00000253023.3 |
UBE2M
|
ubiquitin-conjugating enzyme E2M |
| chr5_-_132113690 | 0.05 |
ENST00000414594.1
|
SEPT8
|
septin 8 |
| chr2_+_149402009 | 0.05 |
ENST00000457184.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr21_-_45079341 | 0.05 |
ENST00000443485.1
ENST00000291560.2 |
HSF2BP
|
heat shock transcription factor 2 binding protein |
| chr7_-_100034060 | 0.05 |
ENST00000292330.2
|
PPP1R35
|
protein phosphatase 1, regulatory subunit 35 |
| chr4_-_16085657 | 0.05 |
ENST00000543373.1
|
PROM1
|
prominin 1 |
| chr17_-_39674668 | 0.05 |
ENST00000393981.3
|
KRT15
|
keratin 15 |
| chr3_+_46618727 | 0.05 |
ENST00000296145.5
|
TDGF1
|
teratocarcinoma-derived growth factor 1 |
| chr17_-_1532106 | 0.05 |
ENST00000301335.5
ENST00000382147.4 |
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr19_-_50143452 | 0.05 |
ENST00000246792.3
|
RRAS
|
related RAS viral (r-ras) oncogene homolog |
| chr12_-_49351148 | 0.05 |
ENST00000398092.4
ENST00000539611.1 |
RP11-302B13.5
ARF3
|
ADP-ribosylation factor 3 ADP-ribosylation factor 3 |
| chr9_-_133814455 | 0.05 |
ENST00000448616.1
|
FIBCD1
|
fibrinogen C domain containing 1 |
| chr14_+_24584508 | 0.05 |
ENST00000559354.1
ENST00000560459.1 ENST00000559593.1 ENST00000396941.4 ENST00000396936.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr1_+_38273818 | 0.05 |
ENST00000373042.4
|
C1orf122
|
chromosome 1 open reading frame 122 |
| chr14_-_71276211 | 0.05 |
ENST00000381250.4
ENST00000555993.2 |
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9 |
| chr17_-_58469474 | 0.05 |
ENST00000300896.4
|
USP32
|
ubiquitin specific peptidase 32 |
| chr16_-_2059797 | 0.05 |
ENST00000563630.1
|
ZNF598
|
zinc finger protein 598 |
| chr9_+_132934835 | 0.05 |
ENST00000372398.3
|
NCS1
|
neuronal calcium sensor 1 |
| chr2_-_74692473 | 0.05 |
ENST00000535045.1
ENST00000409065.1 ENST00000414701.1 ENST00000448666.1 ENST00000233616.4 ENST00000452063.2 |
MOGS
|
mannosyl-oligosaccharide glucosidase |
| chr4_-_16085340 | 0.04 |
ENST00000508167.1
|
PROM1
|
prominin 1 |
| chr9_-_96717654 | 0.04 |
ENST00000253968.6
|
BARX1
|
BARX homeobox 1 |
| chr16_+_89724188 | 0.04 |
ENST00000301031.4
ENST00000566204.1 ENST00000579310.1 |
SPATA33
|
spermatogenesis associated 33 |
| chr1_+_38273988 | 0.04 |
ENST00000446260.2
|
C1orf122
|
chromosome 1 open reading frame 122 |
| chr22_-_24110063 | 0.04 |
ENST00000520222.1
ENST00000401675.3 |
CHCHD10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr20_+_37590942 | 0.04 |
ENST00000373325.2
ENST00000252011.3 ENST00000373323.4 |
DHX35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr16_+_88493879 | 0.04 |
ENST00000565624.1
ENST00000437464.1 |
ZNF469
|
zinc finger protein 469 |
| chr9_+_131873227 | 0.04 |
ENST00000358994.4
ENST00000455292.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr16_+_15528332 | 0.04 |
ENST00000566490.1
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr4_-_105416039 | 0.04 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr1_+_167691191 | 0.04 |
ENST00000392121.3
ENST00000474859.1 |
MPZL1
|
myelin protein zero-like 1 |
| chr22_-_46373004 | 0.04 |
ENST00000339464.4
|
WNT7B
|
wingless-type MMTV integration site family, member 7B |
| chr5_+_49963239 | 0.04 |
ENST00000505554.1
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr20_-_17662705 | 0.04 |
ENST00000455029.2
|
RRBP1
|
ribosome binding protein 1 |
| chr19_+_36203830 | 0.04 |
ENST00000262630.3
|
ZBTB32
|
zinc finger and BTB domain containing 32 |
| chr17_-_40428359 | 0.04 |
ENST00000293328.3
|
STAT5B
|
signal transducer and activator of transcription 5B |
| chr13_+_110959598 | 0.04 |
ENST00000360467.5
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr19_+_38755203 | 0.04 |
ENST00000587090.1
ENST00000454580.3 |
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr15_+_36871806 | 0.04 |
ENST00000566621.1
ENST00000564586.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr1_-_201123586 | 0.04 |
ENST00000414605.2
ENST00000367334.5 ENST00000367332.1 |
TMEM9
|
transmembrane protein 9 |
| chr5_+_443268 | 0.04 |
ENST00000512944.1
|
EXOC3
|
exocyst complex component 3 |
| chr19_-_33793430 | 0.04 |
ENST00000498907.2
|
CEBPA
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr2_-_97536490 | 0.04 |
ENST00000449330.1
|
SEMA4C
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr8_-_74884511 | 0.04 |
ENST00000518127.1
|
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) |
| chr3_-_71114066 | 0.04 |
ENST00000485326.2
|
FOXP1
|
forkhead box P1 |
| chr12_+_51985001 | 0.04 |
ENST00000354534.6
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit |
| chr8_-_134584092 | 0.04 |
ENST00000522652.1
|
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr11_+_67056867 | 0.03 |
ENST00000514166.1
|
ANKRD13D
|
ankyrin repeat domain 13 family, member D |
| chr5_-_140070897 | 0.03 |
ENST00000448240.1
ENST00000438307.2 ENST00000415192.2 ENST00000457527.2 ENST00000307633.3 ENST00000507746.1 ENST00000431330.2 |
HARS
|
histidyl-tRNA synthetase |
| chr1_-_201123546 | 0.03 |
ENST00000435310.1
ENST00000485839.2 ENST00000367330.1 |
TMEM9
|
transmembrane protein 9 |
| chr10_-_131762105 | 0.03 |
ENST00000368648.3
ENST00000355311.5 |
EBF3
|
early B-cell factor 3 |
| chr12_-_51611477 | 0.03 |
ENST00000389243.4
|
POU6F1
|
POU class 6 homeobox 1 |
| chr9_-_131872928 | 0.03 |
ENST00000455830.2
ENST00000393384.3 ENST00000318080.2 |
CRAT
|
carnitine O-acetyltransferase |
| chr17_-_37844267 | 0.03 |
ENST00000579146.1
ENST00000378011.4 ENST00000429199.2 ENST00000300658.4 |
PGAP3
|
post-GPI attachment to proteins 3 |
| chr15_+_92397051 | 0.03 |
ENST00000424469.2
|
SLCO3A1
|
solute carrier organic anion transporter family, member 3A1 |
| chr14_-_59951112 | 0.03 |
ENST00000247194.4
|
L3HYPDH
|
L-3-hydroxyproline dehydratase (trans-) |
| chr22_+_38004832 | 0.03 |
ENST00000405147.3
ENST00000429218.1 ENST00000325180.8 ENST00000337437.4 |
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr16_+_2059872 | 0.03 |
ENST00000567649.1
|
NPW
|
neuropeptide W |
| chr22_+_38004723 | 0.03 |
ENST00000381756.5
|
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr18_-_9614863 | 0.03 |
ENST00000584074.1
|
PPP4R1
|
protein phosphatase 4, regulatory subunit 1 |
| chr1_-_32110467 | 0.03 |
ENST00000440872.2
ENST00000373703.4 |
PEF1
|
penta-EF-hand domain containing 1 |
| chr14_-_24584138 | 0.03 |
ENST00000558280.1
ENST00000561028.1 |
NRL
|
neural retina leucine zipper |
| chr19_-_49622348 | 0.03 |
ENST00000408991.2
|
C19orf73
|
chromosome 19 open reading frame 73 |
| chr19_-_46296011 | 0.03 |
ENST00000377735.3
ENST00000270223.6 |
DMWD
|
dystrophia myotonica, WD repeat containing |
| chr20_-_36156125 | 0.03 |
ENST00000397135.1
ENST00000397137.1 |
BLCAP
|
bladder cancer associated protein |
| chr14_-_51411146 | 0.03 |
ENST00000532462.1
|
PYGL
|
phosphorylase, glycogen, liver |
| chr19_+_36036583 | 0.03 |
ENST00000392205.1
|
TMEM147
|
transmembrane protein 147 |
| chr19_+_38755237 | 0.03 |
ENST00000587516.1
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr11_+_67056755 | 0.03 |
ENST00000511455.2
ENST00000308440.6 |
ANKRD13D
|
ankyrin repeat domain 13 family, member D |
| chr9_-_131790464 | 0.03 |
ENST00000417224.1
ENST00000416629.1 ENST00000372559.1 |
SH3GLB2
|
SH3-domain GRB2-like endophilin B2 |
| chr10_-_16859361 | 0.03 |
ENST00000377921.3
|
RSU1
|
Ras suppressor protein 1 |
| chr1_-_156721502 | 0.03 |
ENST00000357325.5
|
HDGF
|
hepatoma-derived growth factor |
| chr19_+_40697514 | 0.03 |
ENST00000253055.3
|
MAP3K10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr5_-_132112907 | 0.03 |
ENST00000458488.2
|
SEPT8
|
septin 8 |
| chr11_+_133938820 | 0.03 |
ENST00000299106.4
ENST00000529443.2 |
JAM3
|
junctional adhesion molecule 3 |
| chr1_+_43148625 | 0.03 |
ENST00000436427.1
|
YBX1
|
Y box binding protein 1 |
| chr17_-_27044760 | 0.03 |
ENST00000395243.3
|
RAB34
|
RAB34, member RAS oncogene family |
| chr19_+_10197463 | 0.03 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr1_+_110026544 | 0.03 |
ENST00000369870.3
|
ATXN7L2
|
ataxin 7-like 2 |
| chr12_+_96252706 | 0.03 |
ENST00000266735.5
ENST00000553192.1 ENST00000552085.1 |
SNRPF
|
small nuclear ribonucleoprotein polypeptide F |
| chr4_-_7069760 | 0.03 |
ENST00000264954.4
|
GRPEL1
|
GrpE-like 1, mitochondrial (E. coli) |
| chr19_-_5567996 | 0.03 |
ENST00000448587.1
|
TINCR
|
tissue differentiation-inducing non-protein coding RNA |
| chr5_-_443239 | 0.03 |
ENST00000408966.2
|
C5orf55
|
chromosome 5 open reading frame 55 |
| chr11_+_48002076 | 0.03 |
ENST00000418331.2
ENST00000440289.2 |
PTPRJ
|
protein tyrosine phosphatase, receptor type, J |
| chr1_-_38273840 | 0.03 |
ENST00000373044.2
|
YRDC
|
yrdC N(6)-threonylcarbamoyltransferase domain containing |
| chr19_+_38755042 | 0.03 |
ENST00000301244.7
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chrY_+_15815447 | 0.03 |
ENST00000284856.3
|
TMSB4Y
|
thymosin beta 4, Y-linked |
| chr19_-_14228541 | 0.03 |
ENST00000590853.1
ENST00000308677.4 |
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha |
| chr15_-_91475747 | 0.03 |
ENST00000330334.3
ENST00000559898.1 ENST00000394272.3 |
HDDC3
|
HD domain containing 3 |
| chr17_-_30185971 | 0.03 |
ENST00000378634.2
|
COPRS
|
coordinator of PRMT5, differentiation stimulator |
| chr6_-_34113856 | 0.03 |
ENST00000538487.2
|
GRM4
|
glutamate receptor, metabotropic 4 |
| chr20_+_42543504 | 0.03 |
ENST00000341197.4
|
TOX2
|
TOX high mobility group box family member 2 |
| chr12_-_56122426 | 0.03 |
ENST00000551173.1
|
CD63
|
CD63 molecule |
| chr9_-_77643189 | 0.03 |
ENST00000376837.3
|
C9orf41
|
chromosome 9 open reading frame 41 |
| chr1_+_167691185 | 0.03 |
ENST00000359523.2
|
MPZL1
|
myelin protein zero-like 1 |
| chr1_-_202776392 | 0.03 |
ENST00000235790.4
|
KDM5B
|
lysine (K)-specific demethylase 5B |
| chr6_+_73331520 | 0.02 |
ENST00000342056.2
ENST00000355194.4 |
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5 |
| chrX_-_17879356 | 0.02 |
ENST00000331511.1
ENST00000415486.3 ENST00000545871.1 ENST00000451717.1 |
RAI2
|
retinoic acid induced 2 |
| chr14_-_22005018 | 0.02 |
ENST00000546363.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr4_+_41258786 | 0.02 |
ENST00000503431.1
ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr3_+_50126341 | 0.02 |
ENST00000347869.3
ENST00000469838.1 ENST00000404526.2 ENST00000441305.1 |
RBM5
|
RNA binding motif protein 5 |
| chr14_+_24583836 | 0.02 |
ENST00000559115.1
ENST00000558215.1 ENST00000557810.1 ENST00000561375.1 ENST00000446197.3 ENST00000559796.1 ENST00000560713.1 ENST00000560901.1 ENST00000559382.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr11_-_1330834 | 0.02 |
ENST00000525159.1
ENST00000317204.6 ENST00000542915.1 ENST00000527938.1 ENST00000530541.1 ENST00000263646.7 |
TOLLIP
|
toll interacting protein |
| chr22_+_38005033 | 0.02 |
ENST00000447515.1
ENST00000406772.1 ENST00000431745.1 |
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr5_-_145214848 | 0.02 |
ENST00000505416.1
ENST00000334744.4 ENST00000358004.2 ENST00000511435.1 |
PRELID2
|
PRELI domain containing 2 |
| chr1_+_39571026 | 0.02 |
ENST00000524432.1
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr7_+_43152191 | 0.02 |
ENST00000395891.2
|
HECW1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
| chr21_-_47648665 | 0.02 |
ENST00000450351.1
ENST00000522411.1 ENST00000356396.4 ENST00000397728.3 ENST00000457828.2 |
LSS
|
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) |
| chr17_+_6926339 | 0.02 |
ENST00000293805.5
|
BCL6B
|
B-cell CLL/lymphoma 6, member B |
| chr14_-_68283291 | 0.02 |
ENST00000555452.1
ENST00000347230.4 |
ZFYVE26
|
zinc finger, FYVE domain containing 26 |
| chr8_-_74884341 | 0.02 |
ENST00000284811.8
|
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) |
| chr4_-_185458660 | 0.02 |
ENST00000605270.1
|
RP11-326I11.4
|
RP11-326I11.4 |
| chr11_+_133938955 | 0.02 |
ENST00000534549.1
ENST00000441717.3 |
JAM3
|
junctional adhesion molecule 3 |
| chr20_-_36156264 | 0.02 |
ENST00000445723.1
ENST00000414080.1 |
BLCAP
|
bladder cancer associated protein |
| chr14_+_103058948 | 0.02 |
ENST00000262241.6
|
RCOR1
|
REST corepressor 1 |
| chrX_+_133507389 | 0.02 |
ENST00000370800.4
|
PHF6
|
PHD finger protein 6 |
| chr22_-_38480100 | 0.02 |
ENST00000427592.1
|
SLC16A8
|
solute carrier family 16 (monocarboxylate transporter), member 8 |
| chr14_+_29234870 | 0.02 |
ENST00000382535.3
|
FOXG1
|
forkhead box G1 |
| chr2_+_25015968 | 0.02 |
ENST00000380834.2
ENST00000473706.1 |
CENPO
|
centromere protein O |
| chr1_+_93811438 | 0.02 |
ENST00000370272.4
ENST00000370267.1 |
DR1
|
down-regulator of transcription 1, TBP-binding (negative cofactor 2) |
| chr8_-_74884459 | 0.02 |
ENST00000522337.1
|
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) |
| chr2_+_128180842 | 0.02 |
ENST00000402125.2
|
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr9_+_74764278 | 0.02 |
ENST00000238018.4
ENST00000376989.3 |
GDA
|
guanine deaminase |
| chr3_-_71803474 | 0.02 |
ENST00000448225.1
ENST00000496214.2 |
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr1_-_95007193 | 0.02 |
ENST00000370207.4
ENST00000334047.7 |
F3
|
coagulation factor III (thromboplastin, tissue factor) |
| chr2_+_191513587 | 0.02 |
ENST00000416973.1
ENST00000426601.1 |
NAB1
|
NGFI-A binding protein 1 (EGR1 binding protein 1) |
| chr16_-_2014804 | 0.02 |
ENST00000526522.1
ENST00000527302.1 ENST00000529806.1 ENST00000563194.1 ENST00000343262.4 |
RPS2
|
ribosomal protein S2 |
| chrX_-_153141302 | 0.02 |
ENST00000361699.4
ENST00000543994.1 ENST00000370057.3 ENST00000538883.1 ENST00000361981.3 |
L1CAM
|
L1 cell adhesion molecule |
| chr9_+_140500087 | 0.02 |
ENST00000371421.4
|
ARRDC1
|
arrestin domain containing 1 |
| chr8_-_74884482 | 0.02 |
ENST00000520242.1
ENST00000519082.1 |
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) |
| chr19_-_48673465 | 0.02 |
ENST00000598938.1
|
LIG1
|
ligase I, DNA, ATP-dependent |
| chr15_+_90895471 | 0.02 |
ENST00000354377.3
ENST00000379090.5 |
ZNF774
|
zinc finger protein 774 |
| chr9_-_96213828 | 0.02 |
ENST00000423591.1
|
FAM120AOS
|
family with sequence similarity 120A opposite strand |
| chr14_-_22005197 | 0.02 |
ENST00000541965.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr19_-_663277 | 0.02 |
ENST00000292363.5
|
RNF126
|
ring finger protein 126 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB7A_ZBTB7C
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.0 | 0.2 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.2 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.0 | 0.1 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.1 | GO:1903461 | Okazaki fragment processing involved in mitotic DNA replication(GO:1903461) |
| 0.0 | 0.1 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.2 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.0 | 0.1 | GO:0061183 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) positive regulation of dermatome development(GO:0061184) renal vesicle induction(GO:0072034) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:0072737 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.0 | 0.1 | GO:0099640 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.1 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.1 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.0 | 0.0 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.0 | 0.2 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.0 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.0 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) inner medullary collecting duct development(GO:0072061) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.0 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.0 | 0.0 | GO:0036361 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |