Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
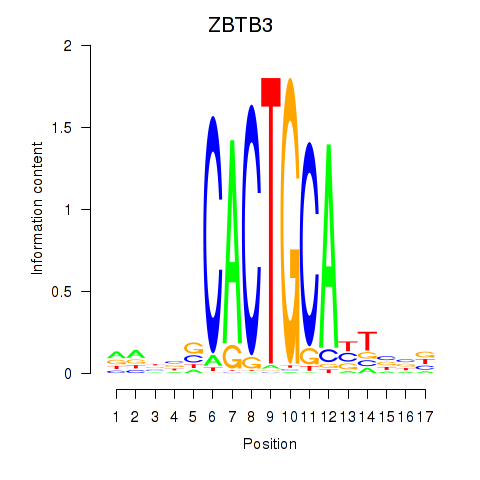
Results for ZBTB3
Z-value: 0.46
Transcription factors associated with ZBTB3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB3
|
ENSG00000185670.7 | zinc finger and BTB domain containing 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB3 | hg19_v2_chr11_-_62521614_62521660 | -0.97 | 3.0e-02 | Click! |
Activity profile of ZBTB3 motif
Sorted Z-values of ZBTB3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_2626988 | 0.30 |
ENST00000509050.1
|
FAM193A
|
family with sequence similarity 193, member A |
| chr5_+_140514782 | 0.27 |
ENST00000231134.5
|
PCDHB5
|
protocadherin beta 5 |
| chrX_-_154563889 | 0.20 |
ENST00000369449.2
ENST00000321926.4 |
CLIC2
|
chloride intracellular channel 2 |
| chr4_-_17783135 | 0.18 |
ENST00000265018.3
|
FAM184B
|
family with sequence similarity 184, member B |
| chr16_+_53164956 | 0.17 |
ENST00000563410.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr14_-_92413727 | 0.16 |
ENST00000267620.10
|
FBLN5
|
fibulin 5 |
| chr19_-_6670128 | 0.16 |
ENST00000245912.3
|
TNFSF14
|
tumor necrosis factor (ligand) superfamily, member 14 |
| chr19_+_39926791 | 0.16 |
ENST00000594990.1
|
SUPT5H
|
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr10_-_105156198 | 0.15 |
ENST00000369815.1
ENST00000309579.3 ENST00000337003.4 |
USMG5
|
up-regulated during skeletal muscle growth 5 homolog (mouse) |
| chr18_+_42260059 | 0.13 |
ENST00000426838.4
|
SETBP1
|
SET binding protein 1 |
| chr7_+_23719749 | 0.12 |
ENST00000409192.3
ENST00000344962.4 ENST00000409653.1 ENST00000409994.3 |
FAM221A
|
family with sequence similarity 221, member A |
| chr8_+_38261880 | 0.11 |
ENST00000527175.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr19_-_19006920 | 0.11 |
ENST00000429504.2
ENST00000427170.2 |
CERS1
|
ceramide synthase 1 |
| chr17_-_5487768 | 0.11 |
ENST00000269280.4
ENST00000345221.3 ENST00000262467.5 |
NLRP1
|
NLR family, pyrin domain containing 1 |
| chr19_-_45657028 | 0.11 |
ENST00000429338.1
ENST00000589776.1 |
NKPD1
|
NTPase, KAP family P-loop domain containing 1 |
| chr4_-_140216948 | 0.10 |
ENST00000265500.4
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr10_+_99332198 | 0.10 |
ENST00000307518.5
ENST00000298808.5 ENST00000370655.1 |
ANKRD2
|
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr3_+_139063372 | 0.10 |
ENST00000478464.1
|
MRPS22
|
mitochondrial ribosomal protein S22 |
| chr12_-_120241187 | 0.10 |
ENST00000392520.2
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr16_+_87636474 | 0.10 |
ENST00000284262.2
|
JPH3
|
junctophilin 3 |
| chr9_+_140500126 | 0.10 |
ENST00000431925.2
ENST00000419386.1 |
ARRDC1
|
arrestin domain containing 1 |
| chr8_+_42196000 | 0.09 |
ENST00000518925.1
ENST00000538005.1 |
POLB
|
polymerase (DNA directed), beta |
| chr16_-_2185899 | 0.09 |
ENST00000262304.4
ENST00000423118.1 |
PKD1
|
polycystic kidney disease 1 (autosomal dominant) |
| chr11_-_82782952 | 0.09 |
ENST00000534141.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr5_+_170288856 | 0.09 |
ENST00000523189.1
|
RANBP17
|
RAN binding protein 17 |
| chr9_+_140500087 | 0.09 |
ENST00000371421.4
|
ARRDC1
|
arrestin domain containing 1 |
| chr1_+_110036728 | 0.09 |
ENST00000369868.3
ENST00000430195.2 |
CYB561D1
|
cytochrome b561 family, member D1 |
| chr1_+_211433275 | 0.08 |
ENST00000367005.4
|
RCOR3
|
REST corepressor 3 |
| chr3_+_139062838 | 0.08 |
ENST00000310776.4
ENST00000465056.1 ENST00000465373.1 |
MRPS22
|
mitochondrial ribosomal protein S22 |
| chr7_+_16793160 | 0.08 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr8_+_144821557 | 0.08 |
ENST00000534398.1
|
FAM83H-AS1
|
FAM83H antisense RNA 1 (head to head) |
| chr19_-_40023450 | 0.08 |
ENST00000326282.4
|
EID2B
|
EP300 interacting inhibitor of differentiation 2B |
| chr9_+_74526532 | 0.08 |
ENST00000486911.2
|
C9orf85
|
chromosome 9 open reading frame 85 |
| chr20_+_62289640 | 0.07 |
ENST00000508582.2
ENST00000360203.5 ENST00000356810.4 |
RTEL1
|
regulator of telomere elongation helicase 1 |
| chr3_-_53878644 | 0.07 |
ENST00000481668.1
ENST00000467802.1 |
CHDH
|
choline dehydrogenase |
| chr2_+_42396472 | 0.07 |
ENST00000318522.5
ENST00000402711.2 |
EML4
|
echinoderm microtubule associated protein like 4 |
| chr1_+_110036699 | 0.07 |
ENST00000496961.1
ENST00000533024.1 ENST00000310611.4 ENST00000527072.1 ENST00000420578.2 ENST00000528785.1 |
CYB561D1
|
cytochrome b561 family, member D1 |
| chr9_-_34637718 | 0.06 |
ENST00000378892.1
ENST00000277010.4 |
SIGMAR1
|
sigma non-opioid intracellular receptor 1 |
| chr4_+_48492269 | 0.06 |
ENST00000327939.4
|
ZAR1
|
zygote arrest 1 |
| chr10_-_118032979 | 0.06 |
ENST00000355422.6
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr14_-_92413353 | 0.06 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr3_-_149688896 | 0.06 |
ENST00000239940.7
|
PFN2
|
profilin 2 |
| chr6_+_30851205 | 0.06 |
ENST00000515881.1
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chrX_+_134166333 | 0.06 |
ENST00000257013.7
|
FAM127A
|
family with sequence similarity 127, member A |
| chr5_+_74807581 | 0.06 |
ENST00000241436.4
ENST00000352007.5 |
POLK
|
polymerase (DNA directed) kappa |
| chr3_-_149688502 | 0.05 |
ENST00000481767.1
ENST00000475518.1 |
PFN2
|
profilin 2 |
| chr9_+_72002837 | 0.05 |
ENST00000377216.3
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chrX_+_11776278 | 0.05 |
ENST00000312196.4
ENST00000337339.2 |
MSL3
|
male-specific lethal 3 homolog (Drosophila) |
| chr16_+_30034655 | 0.05 |
ENST00000300575.2
|
C16orf92
|
chromosome 16 open reading frame 92 |
| chr6_-_31939734 | 0.05 |
ENST00000375356.3
|
DXO
|
decapping exoribonuclease |
| chr6_-_31940065 | 0.05 |
ENST00000375349.3
ENST00000337523.5 |
DXO
|
decapping exoribonuclease |
| chr14_-_92414055 | 0.05 |
ENST00000342058.4
|
FBLN5
|
fibulin 5 |
| chr3_-_149688971 | 0.05 |
ENST00000498307.1
ENST00000489155.1 |
PFN2
|
profilin 2 |
| chr2_-_74570520 | 0.05 |
ENST00000394019.2
ENST00000346834.4 ENST00000359484.4 ENST00000423644.1 ENST00000377634.4 ENST00000436454.1 |
SLC4A5
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 5 |
| chr11_-_82782861 | 0.05 |
ENST00000524635.1
ENST00000526205.1 ENST00000527633.1 ENST00000533486.1 ENST00000533276.2 |
RAB30
|
RAB30, member RAS oncogene family |
| chr19_+_47852538 | 0.05 |
ENST00000328771.4
|
DHX34
|
DEAH (Asp-Glu-Ala-His) box polypeptide 34 |
| chr21_+_45209394 | 0.05 |
ENST00000497547.1
|
RRP1
|
ribosomal RNA processing 1 |
| chr19_-_19006890 | 0.04 |
ENST00000247005.6
|
GDF1
|
growth differentiation factor 1 |
| chr19_+_49496705 | 0.04 |
ENST00000595090.1
|
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr5_+_76506706 | 0.04 |
ENST00000340978.3
ENST00000346042.3 ENST00000264917.5 ENST00000342343.4 ENST00000333194.4 |
PDE8B
|
phosphodiesterase 8B |
| chr1_+_110036674 | 0.04 |
ENST00000393709.3
|
CYB561D1
|
cytochrome b561 family, member D1 |
| chr17_-_39623681 | 0.04 |
ENST00000225899.3
|
KRT32
|
keratin 32 |
| chr16_-_68406161 | 0.04 |
ENST00000568373.1
ENST00000563226.1 |
SMPD3
|
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II) |
| chr5_-_76382989 | 0.04 |
ENST00000511587.1
|
ZBED3
|
zinc finger, BED-type containing 3 |
| chr20_+_54987168 | 0.04 |
ENST00000360314.3
|
CASS4
|
Cas scaffolding protein family member 4 |
| chr12_+_48577366 | 0.03 |
ENST00000316554.3
|
C12orf68
|
chromosome 12 open reading frame 68 |
| chr5_-_94417314 | 0.03 |
ENST00000505208.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr2_+_42396574 | 0.03 |
ENST00000401738.3
|
EML4
|
echinoderm microtubule associated protein like 4 |
| chr5_+_112074029 | 0.03 |
ENST00000512211.2
|
APC
|
adenomatous polyposis coli |
| chr1_-_32801825 | 0.03 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr5_+_75378997 | 0.03 |
ENST00000502798.2
|
SV2C
|
synaptic vesicle glycoprotein 2C |
| chr5_-_94417562 | 0.03 |
ENST00000505465.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr1_-_236046872 | 0.03 |
ENST00000536965.1
|
LYST
|
lysosomal trafficking regulator |
| chr16_+_28986134 | 0.03 |
ENST00000352260.7
|
SPNS1
|
spinster homolog 1 (Drosophila) |
| chrX_+_70364667 | 0.03 |
ENST00000536169.1
ENST00000395855.2 ENST00000374051.3 ENST00000358741.3 |
NLGN3
|
neuroligin 3 |
| chr1_+_160160346 | 0.03 |
ENST00000368078.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr19_+_49496782 | 0.03 |
ENST00000601968.1
ENST00000596837.1 |
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr2_-_40679186 | 0.02 |
ENST00000406785.2
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr21_-_48024986 | 0.02 |
ENST00000291700.4
ENST00000367071.4 |
S100B
|
S100 calcium binding protein B |
| chr19_+_52848659 | 0.02 |
ENST00000327920.8
|
ZNF610
|
zinc finger protein 610 |
| chr4_-_5894777 | 0.02 |
ENST00000324989.7
|
CRMP1
|
collapsin response mediator protein 1 |
| chr3_-_195163803 | 0.02 |
ENST00000326793.6
|
ACAP2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr6_+_44126545 | 0.02 |
ENST00000532171.1
ENST00000398776.1 ENST00000542245.1 |
CAPN11
|
calpain 11 |
| chr11_-_130786400 | 0.02 |
ENST00000265909.4
|
SNX19
|
sorting nexin 19 |
| chr3_-_49131013 | 0.02 |
ENST00000424300.1
|
QRICH1
|
glutamine-rich 1 |
| chr14_-_90085458 | 0.02 |
ENST00000345097.4
ENST00000555855.1 ENST00000555353.1 |
FOXN3
|
forkhead box N3 |
| chr15_-_74659978 | 0.02 |
ENST00000541301.1
ENST00000416978.1 ENST00000268053.6 |
CYP11A1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr1_+_210001309 | 0.02 |
ENST00000491415.2
|
DIEXF
|
digestive organ expansion factor homolog (zebrafish) |
| chr19_+_16187085 | 0.02 |
ENST00000300933.4
|
TPM4
|
tropomyosin 4 |
| chr7_-_38403077 | 0.02 |
ENST00000426402.2
|
TRGV2
|
T cell receptor gamma variable 2 |
| chr9_+_125132803 | 0.01 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr12_+_123949053 | 0.01 |
ENST00000350887.5
|
SNRNP35
|
small nuclear ribonucleoprotein 35kDa (U11/U12) |
| chr17_+_72983674 | 0.01 |
ENST00000337231.5
|
CDR2L
|
cerebellar degeneration-related protein 2-like |
| chr9_+_74526384 | 0.01 |
ENST00000334731.2
ENST00000377031.3 |
C9orf85
|
chromosome 9 open reading frame 85 |
| chr4_+_159236462 | 0.01 |
ENST00000460056.2
|
RXFP1
|
relaxin/insulin-like family peptide receptor 1 |
| chr7_+_100081542 | 0.01 |
ENST00000300179.2
ENST00000423930.1 |
NYAP1
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 1 |
| chr12_-_62586543 | 0.01 |
ENST00000416284.3
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chr10_-_76868866 | 0.01 |
ENST00000607487.1
|
DUSP13
|
dual specificity phosphatase 13 |
| chr7_-_38394118 | 0.01 |
ENST00000390345.2
|
TRGV4
|
T cell receptor gamma variable 4 |
| chr1_+_159141397 | 0.01 |
ENST00000368124.4
ENST00000368125.4 ENST00000416746.1 |
CADM3
|
cell adhesion molecule 3 |
| chr16_+_28986085 | 0.01 |
ENST00000565975.1
ENST00000311008.11 ENST00000323081.8 ENST00000334536.8 |
SPNS1
|
spinster homolog 1 (Drosophila) |
| chr13_+_27998681 | 0.01 |
ENST00000381140.4
|
GTF3A
|
general transcription factor IIIA |
| chr18_+_32073253 | 0.01 |
ENST00000283365.9
ENST00000596745.1 ENST00000315456.6 |
DTNA
|
dystrobrevin, alpha |
| chr1_+_154229547 | 0.01 |
ENST00000428595.1
|
UBAP2L
|
ubiquitin associated protein 2-like |
| chr11_-_18548426 | 0.00 |
ENST00000357193.3
ENST00000536719.1 |
TSG101
|
tumor susceptibility 101 |
| chr3_-_149688655 | 0.00 |
ENST00000461930.1
ENST00000423691.2 ENST00000490975.1 ENST00000461868.1 ENST00000452853.2 |
PFN2
|
profilin 2 |
| chr19_-_49496557 | 0.00 |
ENST00000323798.3
ENST00000541188.1 ENST00000544287.1 ENST00000540532.1 ENST00000263276.6 |
GYS1
|
glycogen synthase 1 (muscle) |
| chr3_+_178276488 | 0.00 |
ENST00000432997.1
ENST00000455865.1 |
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr6_-_46138676 | 0.00 |
ENST00000371383.2
ENST00000230565.3 |
ENPP5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 (putative) |
| chr1_+_65210772 | 0.00 |
ENST00000371072.4
ENST00000294428.3 |
RAVER2
|
ribonucleoprotein, PTB-binding 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0072720 | cellular response to mycotoxin(GO:0036146) response to dithiothreitol(GO:0072720) |
| 0.0 | 0.1 | GO:1904784 | NLRP1 inflammasome complex assembly(GO:1904784) |
| 0.0 | 0.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 0.1 | GO:0072287 | metanephric distal tubule morphogenesis(GO:0072287) |
| 0.0 | 0.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.1 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.1 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.2 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.0 | 0.0 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0002133 | polycystin complex(GO:0002133) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |