Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
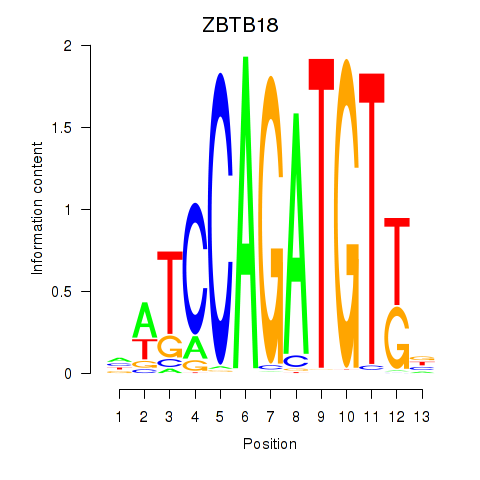
Results for ZBTB18
Z-value: 1.11
Transcription factors associated with ZBTB18
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB18
|
ENSG00000179456.9 | zinc finger and BTB domain containing 18 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB18 | hg19_v2_chr1_+_244214577_244214593 | -0.91 | 8.5e-02 | Click! |
Activity profile of ZBTB18 motif
Sorted Z-values of ZBTB18 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_94577074 | 1.40 |
ENST00000444961.1
ENST00000448882.1 ENST00000557098.1 ENST00000554800.1 ENST00000556544.1 ENST00000298902.5 ENST00000555819.1 ENST00000557634.1 ENST00000555744.1 |
IFI27
|
interferon, alpha-inducible protein 27 |
| chr2_-_7005785 | 1.34 |
ENST00000256722.5
ENST00000404168.1 ENST00000458098.1 |
CMPK2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr7_+_100770328 | 1.18 |
ENST00000223095.4
ENST00000445463.2 |
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr17_+_48638371 | 1.12 |
ENST00000360761.4
ENST00000352832.5 ENST00000354983.4 |
CACNA1G
|
calcium channel, voltage-dependent, T type, alpha 1G subunit |
| chr11_+_4510109 | 0.96 |
ENST00000307632.3
|
OR52K1
|
olfactory receptor, family 52, subfamily K, member 1 |
| chr9_+_100174344 | 0.73 |
ENST00000422139.2
|
TDRD7
|
tudor domain containing 7 |
| chr9_+_100174232 | 0.73 |
ENST00000355295.4
|
TDRD7
|
tudor domain containing 7 |
| chr10_+_90562705 | 0.72 |
ENST00000539337.1
|
LIPM
|
lipase, family member M |
| chr14_-_91294472 | 0.70 |
ENST00000555975.1
|
CTD-3035D6.2
|
CTD-3035D6.2 |
| chr2_-_152146385 | 0.69 |
ENST00000414946.1
ENST00000243346.5 |
NMI
|
N-myc (and STAT) interactor |
| chr11_+_4470525 | 0.69 |
ENST00000325719.4
|
OR52K2
|
olfactory receptor, family 52, subfamily K, member 2 |
| chr12_+_112563303 | 0.64 |
ENST00000412615.2
|
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chr17_+_77020325 | 0.61 |
ENST00000311661.4
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chrX_-_153141302 | 0.60 |
ENST00000361699.4
ENST00000543994.1 ENST00000370057.3 ENST00000538883.1 ENST00000361981.3 |
L1CAM
|
L1 cell adhesion molecule |
| chr17_+_77020224 | 0.60 |
ENST00000339142.2
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr17_+_77020146 | 0.59 |
ENST00000579760.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr12_-_54785074 | 0.59 |
ENST00000338010.5
ENST00000550774.1 |
ZNF385A
|
zinc finger protein 385A |
| chr12_-_54785054 | 0.57 |
ENST00000352268.6
ENST00000549962.1 |
ZNF385A
|
zinc finger protein 385A |
| chr12_+_112563335 | 0.56 |
ENST00000549358.1
ENST00000257604.5 ENST00000548092.1 ENST00000552896.1 |
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chr14_+_64971438 | 0.52 |
ENST00000555321.1
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr11_-_5537920 | 0.48 |
ENST00000380184.1
|
UBQLNL
|
ubiquilin-like |
| chr8_+_38965048 | 0.47 |
ENST00000399831.3
ENST00000437682.2 ENST00000519315.1 ENST00000379907.4 ENST00000522506.1 |
ADAM32
|
ADAM metallopeptidase domain 32 |
| chr19_+_41119323 | 0.46 |
ENST00000599724.1
ENST00000597071.1 ENST00000243562.9 |
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr4_+_75311019 | 0.45 |
ENST00000502307.1
|
AREG
|
amphiregulin |
| chr19_+_54135310 | 0.43 |
ENST00000376650.1
|
DPRX
|
divergent-paired related homeobox |
| chrX_-_19688475 | 0.43 |
ENST00000541422.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr2_-_73511407 | 0.42 |
ENST00000520530.2
|
FBXO41
|
F-box protein 41 |
| chr11_-_119252425 | 0.42 |
ENST00000260187.2
|
USP2
|
ubiquitin specific peptidase 2 |
| chr15_-_74501360 | 0.42 |
ENST00000323940.5
|
STRA6
|
stimulated by retinoic acid 6 |
| chr12_-_104443890 | 0.41 |
ENST00000547583.1
ENST00000360814.4 ENST00000546851.1 |
GLT8D2
|
glycosyltransferase 8 domain containing 2 |
| chr11_+_76494253 | 0.40 |
ENST00000333090.4
|
TSKU
|
tsukushi, small leucine rich proteoglycan |
| chr10_+_90562487 | 0.40 |
ENST00000404743.4
|
LIPM
|
lipase, family member M |
| chr8_-_49833978 | 0.40 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr12_+_27619743 | 0.40 |
ENST00000298876.4
ENST00000416383.1 |
SMCO2
|
single-pass membrane protein with coiled-coil domains 2 |
| chr2_-_163099885 | 0.39 |
ENST00000443424.1
|
FAP
|
fibroblast activation protein, alpha |
| chr17_+_64961026 | 0.38 |
ENST00000262138.3
|
CACNG4
|
calcium channel, voltage-dependent, gamma subunit 4 |
| chr11_-_119252359 | 0.38 |
ENST00000455332.2
|
USP2
|
ubiquitin specific peptidase 2 |
| chr15_-_74501310 | 0.38 |
ENST00000423167.2
ENST00000432245.2 |
STRA6
|
stimulated by retinoic acid 6 |
| chr11_-_62323702 | 0.37 |
ENST00000530285.1
|
AHNAK
|
AHNAK nucleoprotein |
| chr4_+_75310851 | 0.37 |
ENST00000395748.3
ENST00000264487.2 |
AREG
|
amphiregulin |
| chr1_+_19967014 | 0.37 |
ENST00000428975.1
|
NBL1
|
neuroblastoma 1, DAN family BMP antagonist |
| chr8_-_73793975 | 0.36 |
ENST00000523881.1
|
RP11-1145L24.1
|
RP11-1145L24.1 |
| chr7_+_105172612 | 0.35 |
ENST00000493041.1
|
RINT1
|
RAD50 interactor 1 |
| chr18_+_3448455 | 0.33 |
ENST00000549780.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr17_+_32582293 | 0.32 |
ENST00000580907.1
ENST00000225831.4 |
CCL2
|
chemokine (C-C motif) ligand 2 |
| chrX_-_153141434 | 0.32 |
ENST00000407935.2
ENST00000439496.1 |
L1CAM
|
L1 cell adhesion molecule |
| chr2_-_163100045 | 0.31 |
ENST00000188790.4
|
FAP
|
fibroblast activation protein, alpha |
| chr4_+_75480629 | 0.31 |
ENST00000380846.3
|
AREGB
|
amphiregulin B |
| chr2_+_177053307 | 0.31 |
ENST00000331462.4
|
HOXD1
|
homeobox D1 |
| chr3_-_160167301 | 0.30 |
ENST00000494486.1
|
TRIM59
|
tripartite motif containing 59 |
| chr17_-_41174424 | 0.30 |
ENST00000355653.3
|
VAT1
|
vesicle amine transport 1 |
| chr2_-_69064324 | 0.30 |
ENST00000415448.1
|
AC097495.3
|
AC097495.3 |
| chr15_+_67418047 | 0.29 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr14_-_64971288 | 0.29 |
ENST00000394715.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chr8_-_49834299 | 0.28 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2 |
| chr15_-_74043816 | 0.27 |
ENST00000379822.4
|
C15orf59
|
chromosome 15 open reading frame 59 |
| chr9_+_127023704 | 0.27 |
ENST00000373596.1
ENST00000425237.1 |
NEK6
|
NIMA-related kinase 6 |
| chr4_+_144312659 | 0.27 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr11_-_34379546 | 0.26 |
ENST00000435224.2
|
ABTB2
|
ankyrin repeat and BTB (POZ) domain containing 2 |
| chrX_+_48242863 | 0.26 |
ENST00000376886.2
ENST00000375517.3 |
SSX4
|
synovial sarcoma, X breakpoint 4 |
| chr19_+_39903185 | 0.26 |
ENST00000409794.3
|
PLEKHG2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr6_-_16761678 | 0.25 |
ENST00000244769.4
ENST00000436367.1 |
ATXN1
|
ataxin 1 |
| chr1_-_217262969 | 0.24 |
ENST00000361525.3
|
ESRRG
|
estrogen-related receptor gamma |
| chr2_+_189839046 | 0.24 |
ENST00000304636.3
ENST00000317840.5 |
COL3A1
|
collagen, type III, alpha 1 |
| chr11_-_128894053 | 0.22 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr2_+_201994042 | 0.21 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr10_+_11206925 | 0.21 |
ENST00000354440.2
ENST00000315874.4 ENST00000427450.1 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr1_+_78354330 | 0.21 |
ENST00000440324.1
|
NEXN
|
nexilin (F actin binding protein) |
| chr17_+_32597232 | 0.21 |
ENST00000378569.2
ENST00000200307.4 ENST00000394627.1 ENST00000394630.3 |
CCL7
|
chemokine (C-C motif) ligand 7 |
| chr7_-_122339162 | 0.20 |
ENST00000340112.2
|
RNF133
|
ring finger protein 133 |
| chr1_-_113160826 | 0.20 |
ENST00000538187.1
ENST00000369664.1 |
ST7L
|
suppression of tumorigenicity 7 like |
| chr5_-_142784888 | 0.20 |
ENST00000514699.1
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr11_+_32154200 | 0.20 |
ENST00000525133.1
|
RP1-65P5.5
|
RP1-65P5.5 |
| chr3_-_37216055 | 0.20 |
ENST00000336686.4
|
LRRFIP2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr5_+_159895275 | 0.19 |
ENST00000517927.1
|
MIR146A
|
microRNA 146a |
| chrX_-_77225135 | 0.19 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr9_+_131683174 | 0.19 |
ENST00000372592.3
ENST00000428610.1 |
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr6_-_74363803 | 0.19 |
ENST00000355773.5
|
SLC17A5
|
solute carrier family 17 (acidic sugar transporter), member 5 |
| chr6_-_163148700 | 0.19 |
ENST00000366894.1
ENST00000338468.3 |
PARK2
|
parkin RBR E3 ubiquitin protein ligase |
| chr12_+_2162447 | 0.19 |
ENST00000335762.5
ENST00000399655.1 |
CACNA1C
|
calcium channel, voltage-dependent, L type, alpha 1C subunit |
| chr8_+_38662960 | 0.18 |
ENST00000524193.1
|
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr19_-_36001386 | 0.18 |
ENST00000461300.1
|
DMKN
|
dermokine |
| chr12_-_48119340 | 0.18 |
ENST00000229003.3
|
ENDOU
|
endonuclease, polyU-specific |
| chr2_-_65659762 | 0.17 |
ENST00000440972.1
|
SPRED2
|
sprouty-related, EVH1 domain containing 2 |
| chr2_-_145278475 | 0.17 |
ENST00000558170.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr2_-_85829811 | 0.17 |
ENST00000306353.3
|
TMEM150A
|
transmembrane protein 150A |
| chr17_+_47653178 | 0.17 |
ENST00000328741.5
|
NXPH3
|
neurexophilin 3 |
| chr10_-_104913367 | 0.17 |
ENST00000423468.2
|
NT5C2
|
5'-nucleotidase, cytosolic II |
| chr4_+_47487285 | 0.17 |
ENST00000273859.3
ENST00000504445.1 |
ATP10D
|
ATPase, class V, type 10D |
| chr10_-_95241951 | 0.16 |
ENST00000358334.5
ENST00000359263.4 ENST00000371488.3 |
MYOF
|
myoferlin |
| chr3_+_121311966 | 0.16 |
ENST00000338040.4
|
FBXO40
|
F-box protein 40 |
| chr19_+_9251052 | 0.16 |
ENST00000247956.6
ENST00000360385.3 |
ZNF317
|
zinc finger protein 317 |
| chr3_-_52273098 | 0.16 |
ENST00000499914.2
ENST00000305533.5 ENST00000597542.1 |
TWF2
TLR9
|
twinfilin actin-binding protein 2 toll-like receptor 9 |
| chr6_-_127663543 | 0.16 |
ENST00000531582.1
|
ECHDC1
|
enoyl CoA hydratase domain containing 1 |
| chrX_-_48056199 | 0.15 |
ENST00000311798.1
ENST00000347757.1 |
SSX5
|
synovial sarcoma, X breakpoint 5 |
| chr10_-_95242044 | 0.15 |
ENST00000371501.4
ENST00000371502.4 ENST00000371489.1 |
MYOF
|
myoferlin |
| chr1_+_26737253 | 0.15 |
ENST00000326279.6
|
LIN28A
|
lin-28 homolog A (C. elegans) |
| chr19_+_50936142 | 0.15 |
ENST00000357701.5
|
MYBPC2
|
myosin binding protein C, fast type |
| chr9_+_87283430 | 0.15 |
ENST00000376214.1
ENST00000376213.1 |
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr12_-_102874102 | 0.15 |
ENST00000392905.2
|
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
| chr7_+_120969045 | 0.14 |
ENST00000222462.2
|
WNT16
|
wingless-type MMTV integration site family, member 16 |
| chr13_-_33780133 | 0.14 |
ENST00000399365.3
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr6_+_160542821 | 0.14 |
ENST00000366963.4
|
SLC22A1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr18_+_6834472 | 0.14 |
ENST00000581099.1
ENST00000419673.2 ENST00000531294.1 |
ARHGAP28
|
Rho GTPase activating protein 28 |
| chrX_+_150151824 | 0.14 |
ENST00000455596.1
ENST00000448905.2 |
HMGB3
|
high mobility group box 3 |
| chr5_-_142784101 | 0.13 |
ENST00000503201.1
ENST00000502892.1 |
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr12_-_48119301 | 0.13 |
ENST00000545824.2
ENST00000422538.3 |
ENDOU
|
endonuclease, polyU-specific |
| chr1_+_26737292 | 0.13 |
ENST00000254231.4
|
LIN28A
|
lin-28 homolog A (C. elegans) |
| chr3_-_111314230 | 0.13 |
ENST00000317012.4
|
ZBED2
|
zinc finger, BED-type containing 2 |
| chr2_-_74692473 | 0.13 |
ENST00000535045.1
ENST00000409065.1 ENST00000414701.1 ENST00000448666.1 ENST00000233616.4 ENST00000452063.2 |
MOGS
|
mannosyl-oligosaccharide glucosidase |
| chr3_+_183894566 | 0.12 |
ENST00000439647.1
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit |
| chrX_-_153141783 | 0.12 |
ENST00000458029.1
|
L1CAM
|
L1 cell adhesion molecule |
| chr19_+_42806250 | 0.12 |
ENST00000598490.1
ENST00000341747.3 |
PRR19
|
proline rich 19 |
| chr2_-_85829780 | 0.12 |
ENST00000334462.5
|
TMEM150A
|
transmembrane protein 150A |
| chr5_+_140782351 | 0.12 |
ENST00000573521.1
|
PCDHGA9
|
protocadherin gamma subfamily A, 9 |
| chr12_+_57854274 | 0.12 |
ENST00000528432.1
|
GLI1
|
GLI family zinc finger 1 |
| chr16_-_18470696 | 0.12 |
ENST00000427999.2
|
RP11-1212A22.4
|
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr3_-_105588231 | 0.11 |
ENST00000545639.1
ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr10_+_105253661 | 0.11 |
ENST00000369780.4
|
NEURL
|
neuralized E3 ubiquitin protein ligase 1 |
| chr11_+_65405556 | 0.11 |
ENST00000534313.1
ENST00000533361.1 ENST00000526137.1 |
SIPA1
|
signal-induced proliferation-associated 1 |
| chr12_-_54121261 | 0.11 |
ENST00000549784.1
ENST00000262059.4 |
CALCOCO1
|
calcium binding and coiled-coil domain 1 |
| chr15_-_31453359 | 0.11 |
ENST00000542188.1
|
TRPM1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr12_-_16762971 | 0.11 |
ENST00000540590.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr16_-_67185117 | 0.11 |
ENST00000449549.3
|
B3GNT9
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9 |
| chr1_-_32210275 | 0.10 |
ENST00000440175.2
|
BAI2
|
brain-specific angiogenesis inhibitor 2 |
| chr7_-_135433534 | 0.10 |
ENST00000338588.3
|
FAM180A
|
family with sequence similarity 180, member A |
| chr1_-_92951607 | 0.10 |
ENST00000427103.1
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr20_+_30467600 | 0.10 |
ENST00000375934.4
ENST00000375922.4 |
TTLL9
|
tubulin tyrosine ligase-like family, member 9 |
| chr6_+_163148973 | 0.10 |
ENST00000366888.2
|
PACRG
|
PARK2 co-regulated |
| chr8_-_29592736 | 0.10 |
ENST00000518623.1
|
LINC00589
|
long intergenic non-protein coding RNA 589 |
| chr10_+_97733786 | 0.10 |
ENST00000371198.2
|
CC2D2B
|
coiled-coil and C2 domain containing 2B |
| chr5_+_140792614 | 0.09 |
ENST00000398610.2
|
PCDHGA10
|
protocadherin gamma subfamily A, 10 |
| chr12_-_16762802 | 0.09 |
ENST00000534946.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr7_+_95115210 | 0.09 |
ENST00000428113.1
ENST00000325885.5 |
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr7_+_90225796 | 0.09 |
ENST00000380050.3
|
CDK14
|
cyclin-dependent kinase 14 |
| chr1_-_2458026 | 0.09 |
ENST00000435556.3
ENST00000378466.3 |
PANK4
|
pantothenate kinase 4 |
| chr19_-_36001113 | 0.09 |
ENST00000434389.1
|
DMKN
|
dermokine |
| chr1_-_217262933 | 0.08 |
ENST00000359162.2
|
ESRRG
|
estrogen-related receptor gamma |
| chr6_-_163148780 | 0.08 |
ENST00000366892.1
ENST00000366898.1 ENST00000366897.1 ENST00000366896.1 |
PARK2
|
parkin RBR E3 ubiquitin protein ligase |
| chr9_-_13165457 | 0.08 |
ENST00000542239.1
ENST00000538841.1 ENST00000433359.2 |
MPDZ
|
multiple PDZ domain protein |
| chr8_+_24241789 | 0.07 |
ENST00000256412.4
ENST00000538205.1 |
ADAMDEC1
|
ADAM-like, decysin 1 |
| chr15_+_51669444 | 0.07 |
ENST00000396399.2
|
GLDN
|
gliomedin |
| chr8_+_104831472 | 0.07 |
ENST00000262231.10
ENST00000507740.1 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr1_+_38022572 | 0.06 |
ENST00000541606.1
|
DNALI1
|
dynein, axonemal, light intermediate chain 1 |
| chr14_-_61748550 | 0.06 |
ENST00000555868.1
|
TMEM30B
|
transmembrane protein 30B |
| chr14_-_101014435 | 0.06 |
ENST00000554356.1
|
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr16_-_49698136 | 0.06 |
ENST00000535559.1
|
ZNF423
|
zinc finger protein 423 |
| chr8_+_24241969 | 0.06 |
ENST00000522298.1
|
ADAMDEC1
|
ADAM-like, decysin 1 |
| chr10_-_48050538 | 0.06 |
ENST00000420079.2
|
ASAH2C
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2C |
| chrX_+_103031421 | 0.05 |
ENST00000433491.1
ENST00000418604.1 ENST00000443502.1 |
PLP1
|
proteolipid protein 1 |
| chr7_-_154863264 | 0.05 |
ENST00000395731.2
ENST00000543018.1 |
HTR5A-AS1
|
HTR5A antisense RNA 1 |
| chr11_+_128562372 | 0.05 |
ENST00000344954.6
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr17_-_62050278 | 0.05 |
ENST00000578147.1
ENST00000435607.1 |
SCN4A
|
sodium channel, voltage-gated, type IV, alpha subunit |
| chr10_-_7829909 | 0.05 |
ENST00000379562.4
ENST00000543003.1 ENST00000535925.1 |
KIN
|
KIN, antigenic determinant of recA protein homolog (mouse) |
| chr3_-_178789220 | 0.05 |
ENST00000414084.1
|
ZMAT3
|
zinc finger, matrin-type 3 |
| chr3_+_183894737 | 0.05 |
ENST00000432591.1
ENST00000431779.1 |
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit |
| chr12_-_68845165 | 0.04 |
ENST00000360485.3
ENST00000441255.2 |
RP11-81H14.2
|
RP11-81H14.2 |
| chr13_+_113699029 | 0.04 |
ENST00000423251.1
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr12_-_50222187 | 0.04 |
ENST00000335999.6
|
NCKAP5L
|
NCK-associated protein 5-like |
| chr1_+_186265399 | 0.03 |
ENST00000367486.3
ENST00000367484.3 ENST00000533951.1 ENST00000367482.4 ENST00000367483.4 ENST00000367485.4 ENST00000445192.2 |
PRG4
|
proteoglycan 4 |
| chr1_-_151882031 | 0.03 |
ENST00000489410.1
|
THEM4
|
thioesterase superfamily member 4 |
| chr2_+_152214098 | 0.03 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr2_-_85829496 | 0.03 |
ENST00000409668.1
|
TMEM150A
|
transmembrane protein 150A |
| chr9_+_125137565 | 0.03 |
ENST00000373698.5
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr17_+_19912640 | 0.03 |
ENST00000395527.4
ENST00000583482.2 ENST00000583528.1 ENST00000583463.1 |
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr1_+_53527854 | 0.02 |
ENST00000371500.3
ENST00000395871.2 ENST00000312553.5 |
PODN
|
podocan |
| chr3_+_100211412 | 0.02 |
ENST00000323523.4
ENST00000403410.1 ENST00000449609.1 |
TMEM45A
|
transmembrane protein 45A |
| chr12_+_26126681 | 0.02 |
ENST00000542865.1
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr11_+_4673716 | 0.02 |
ENST00000530215.1
|
OR51E1
|
olfactory receptor, family 51, subfamily E, member 1 |
| chr11_+_5509915 | 0.02 |
ENST00000322641.5
|
OR52D1
|
olfactory receptor, family 52, subfamily D, member 1 |
| chr22_-_37213554 | 0.02 |
ENST00000443735.1
|
PVALB
|
parvalbumin |
| chr7_-_47578840 | 0.01 |
ENST00000450444.1
|
TNS3
|
tensin 3 |
| chr3_+_124303512 | 0.01 |
ENST00000454902.1
|
KALRN
|
kalirin, RhoGEF kinase |
| chr19_+_11649532 | 0.01 |
ENST00000252456.2
ENST00000592923.1 ENST00000535659.2 |
CNN1
|
calponin 1, basic, smooth muscle |
| chrX_+_9880590 | 0.01 |
ENST00000452575.1
|
SHROOM2
|
shroom family member 2 |
| chrX_+_103031758 | 0.01 |
ENST00000303958.2
ENST00000361621.2 |
PLP1
|
proteolipid protein 1 |
| chr9_+_273038 | 0.01 |
ENST00000487230.1
ENST00000469391.1 |
DOCK8
|
dedicator of cytokinesis 8 |
| chr3_+_124303539 | 0.01 |
ENST00000428018.2
|
KALRN
|
kalirin, RhoGEF kinase |
| chr1_-_247275719 | 0.01 |
ENST00000408893.2
|
C1orf229
|
chromosome 1 open reading frame 229 |
| chr1_+_38022513 | 0.01 |
ENST00000296218.7
|
DNALI1
|
dynein, axonemal, light intermediate chain 1 |
| chr10_+_7830125 | 0.01 |
ENST00000335698.4
ENST00000541227.1 |
ATP5C1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chrX_+_150151752 | 0.01 |
ENST00000325307.7
|
HMGB3
|
high mobility group box 3 |
| chr1_+_37947257 | 0.01 |
ENST00000471012.1
|
ZC3H12A
|
zinc finger CCCH-type containing 12A |
| chr2_+_20650796 | 0.00 |
ENST00000448241.1
|
AC023137.2
|
AC023137.2 |
| chr19_+_50084561 | 0.00 |
ENST00000246794.5
|
PRRG2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr22_+_42372970 | 0.00 |
ENST00000291236.11
|
SEPT3
|
septin 3 |
| chr3_+_159570722 | 0.00 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr8_+_1772132 | 0.00 |
ENST00000349830.3
ENST00000520359.1 ENST00000518288.1 ENST00000398560.1 |
ARHGEF10
|
Rho guanine nucleotide exchange factor (GEF) 10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB18
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.3 | 1.3 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.2 | 0.7 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.2 | 1.8 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.2 | 1.2 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.2 | 1.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 | 0.7 | GO:0010716 | negative regulation of extracellular matrix disassembly(GO:0010716) |
| 0.1 | 0.8 | GO:0045354 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.1 | 0.8 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.1 | 0.3 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.1 | 0.8 | GO:0060751 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.1 | 0.4 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 0.4 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.0 | 0.2 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) |
| 0.0 | 0.8 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.3 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 1.6 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.1 | GO:1904073 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.0 | 0.2 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 1.4 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.2 | GO:2000501 | regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.0 | 0.1 | GO:0060032 | notochord regression(GO:0060032) |
| 0.0 | 0.3 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.2 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.0 | 0.1 | GO:0099551 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.0 | 0.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 1.4 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.0 | 0.2 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.2 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 1.2 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.0 | 0.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 1.0 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.1 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.0 | 0.5 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.2 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.2 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.7 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.3 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 1.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 1.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 1.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 1.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.3 | 1.3 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 1.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.2 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.1 | 0.3 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.2 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.3 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.2 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.0 | 0.3 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 1.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.4 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 1.4 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.3 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.1 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.1 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.5 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 1.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.3 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.8 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.6 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.7 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 1.8 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |