Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for ZBTB16
Z-value: 0.52
Transcription factors associated with ZBTB16
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB16
|
ENSG00000109906.9 | zinc finger and BTB domain containing 16 |
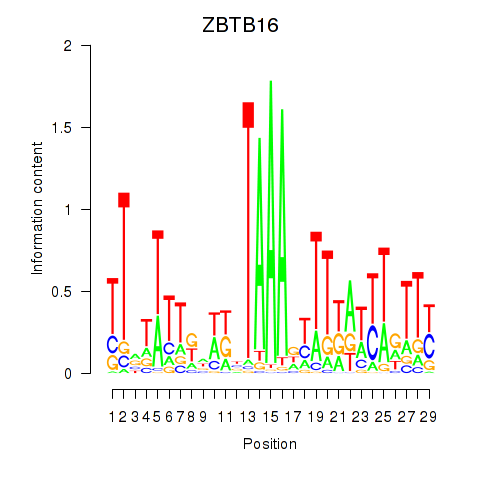
Activity profile of ZBTB16 motif
Sorted Z-values of ZBTB16 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_71533055 | 0.66 |
ENST00000552128.1
|
TSPAN8
|
tetraspanin 8 |
| chr2_-_228244013 | 0.61 |
ENST00000304568.3
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr10_+_62538089 | 0.52 |
ENST00000519078.2
ENST00000395284.3 ENST00000316629.4 |
CDK1
|
cyclin-dependent kinase 1 |
| chr6_-_47445214 | 0.49 |
ENST00000604014.1
|
RP11-385F7.1
|
RP11-385F7.1 |
| chr10_-_98347063 | 0.45 |
ENST00000443638.1
|
TM9SF3
|
transmembrane 9 superfamily member 3 |
| chr10_+_104614008 | 0.40 |
ENST00000369883.3
|
C10orf32
|
chromosome 10 open reading frame 32 |
| chr19_+_42041860 | 0.37 |
ENST00000483481.2
ENST00000494375.2 |
AC006129.4
|
AC006129.4 |
| chr8_-_95229531 | 0.37 |
ENST00000450165.2
|
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr1_+_241695670 | 0.33 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr17_+_21729593 | 0.30 |
ENST00000581769.1
ENST00000584755.1 |
UBBP4
|
ubiquitin B pseudogene 4 |
| chr7_-_16872932 | 0.28 |
ENST00000419572.2
ENST00000412973.1 |
AGR2
|
anterior gradient 2 |
| chr17_+_46908350 | 0.27 |
ENST00000258947.3
ENST00000509507.1 ENST00000448105.2 ENST00000570513.1 ENST00000509415.1 ENST00000513119.1 ENST00000416445.2 ENST00000508679.1 ENST00000505071.1 |
CALCOCO2
|
calcium binding and coiled-coil domain 2 |
| chr9_-_116837249 | 0.26 |
ENST00000466610.2
|
AMBP
|
alpha-1-microglobulin/bikunin precursor |
| chr18_+_6256746 | 0.26 |
ENST00000578427.1
|
RP11-760N9.1
|
RP11-760N9.1 |
| chr3_-_15382875 | 0.23 |
ENST00000408919.3
|
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated) |
| chr4_-_159094194 | 0.23 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chr3_-_93692781 | 0.23 |
ENST00000394236.3
|
PROS1
|
protein S (alpha) |
| chr18_-_52626622 | 0.21 |
ENST00000591504.1
|
CCDC68
|
coiled-coil domain containing 68 |
| chr15_-_54051831 | 0.21 |
ENST00000557913.1
ENST00000360509.5 |
WDR72
|
WD repeat domain 72 |
| chr19_-_14992264 | 0.21 |
ENST00000327462.2
|
OR7A17
|
olfactory receptor, family 7, subfamily A, member 17 |
| chr18_+_3466248 | 0.20 |
ENST00000581029.1
ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4
|
RP11-838N2.4 |
| chr3_+_148415571 | 0.20 |
ENST00000497524.1
ENST00000349243.3 ENST00000542281.1 ENST00000418473.2 ENST00000404754.2 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr1_+_76251912 | 0.19 |
ENST00000370826.3
|
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit |
| chr15_-_54025300 | 0.19 |
ENST00000559418.1
|
WDR72
|
WD repeat domain 72 |
| chrX_+_118602363 | 0.19 |
ENST00000317881.8
|
SLC25A5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr8_+_66619277 | 0.19 |
ENST00000521247.2
ENST00000527155.1 |
MTFR1
|
mitochondrial fission regulator 1 |
| chr3_+_160394940 | 0.19 |
ENST00000320767.2
|
ARL14
|
ADP-ribosylation factor-like 14 |
| chr16_-_20817753 | 0.19 |
ENST00000389345.5
ENST00000300005.3 ENST00000357967.4 ENST00000569729.1 |
ERI2
|
ERI1 exoribonuclease family member 2 |
| chr1_+_207277632 | 0.18 |
ENST00000421786.1
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr3_-_165555200 | 0.18 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr12_+_133657461 | 0.18 |
ENST00000412146.2
ENST00000544426.1 ENST00000440984.2 ENST00000319849.3 ENST00000440550.2 |
ZNF140
|
zinc finger protein 140 |
| chr14_-_45722360 | 0.18 |
ENST00000451174.1
|
MIS18BP1
|
MIS18 binding protein 1 |
| chr16_-_46649905 | 0.18 |
ENST00000569702.1
|
SHCBP1
|
SHC SH2-domain binding protein 1 |
| chr3_+_44840679 | 0.17 |
ENST00000425755.1
|
KIF15
|
kinesin family member 15 |
| chr22_+_38203898 | 0.17 |
ENST00000323205.6
ENST00000248924.6 ENST00000445195.1 |
GCAT
|
glycine C-acetyltransferase |
| chr17_-_74351010 | 0.17 |
ENST00000435555.2
|
PRPSAP1
|
phosphoribosyl pyrophosphate synthetase-associated protein 1 |
| chr10_+_104613980 | 0.17 |
ENST00000339834.5
|
C10orf32
|
chromosome 10 open reading frame 32 |
| chr18_+_11857439 | 0.17 |
ENST00000602628.1
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr12_+_20968608 | 0.16 |
ENST00000381541.3
ENST00000540229.1 ENST00000553473.1 ENST00000554957.1 |
LST3
SLCO1B3
SLCO1B7
|
Putative solute carrier organic anion transporter family member 1B7; Uncharacterized protein solute carrier organic anion transporter family, member 1B3 solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr15_+_66585555 | 0.16 |
ENST00000319194.5
ENST00000525134.2 ENST00000441424.2 |
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr1_+_156095951 | 0.16 |
ENST00000448611.2
ENST00000368297.1 |
LMNA
|
lamin A/C |
| chr12_+_97306295 | 0.16 |
ENST00000457368.2
|
NEDD1
|
neural precursor cell expressed, developmentally down-regulated 1 |
| chr5_+_180682720 | 0.15 |
ENST00000599439.1
|
AC008443.1
|
CDNA: FLJ23158 fis, clone LNG09623; Uncharacterized protein |
| chr4_-_38784572 | 0.15 |
ENST00000308973.4
ENST00000361424.2 |
TLR10
|
toll-like receptor 10 |
| chr22_-_30901637 | 0.15 |
ENST00000381982.3
ENST00000255858.7 ENST00000540456.1 ENST00000392772.2 |
SEC14L4
|
SEC14-like 4 (S. cerevisiae) |
| chr15_+_69365272 | 0.15 |
ENST00000559914.1
ENST00000558369.1 |
LINC00277
|
long intergenic non-protein coding RNA 277 |
| chr5_+_68860949 | 0.15 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr12_-_74686314 | 0.15 |
ENST00000551210.1
ENST00000515416.2 ENST00000549905.1 |
RP11-81H3.2
|
RP11-81H3.2 |
| chr17_-_46623441 | 0.15 |
ENST00000330070.4
|
HOXB2
|
homeobox B2 |
| chr12_-_68696652 | 0.14 |
ENST00000539972.1
|
MDM1
|
Mdm1 nuclear protein homolog (mouse) |
| chr2_+_200472779 | 0.14 |
ENST00000427045.1
ENST00000419243.1 |
AC093590.1
|
AC093590.1 |
| chr7_-_130066571 | 0.14 |
ENST00000492389.1
|
CEP41
|
centrosomal protein 41kDa |
| chr3_+_179065474 | 0.14 |
ENST00000471841.1
ENST00000280653.7 |
MFN1
|
mitofusin 1 |
| chr12_+_85430110 | 0.14 |
ENST00000393212.3
ENST00000393217.2 |
LRRIQ1
|
leucine-rich repeats and IQ motif containing 1 |
| chr22_-_38239808 | 0.14 |
ENST00000406423.1
ENST00000424350.1 ENST00000458278.2 |
ANKRD54
|
ankyrin repeat domain 54 |
| chr12_+_58166726 | 0.14 |
ENST00000546504.1
|
RP11-571M6.15
|
Uncharacterized protein |
| chr7_-_104909435 | 0.13 |
ENST00000357311.3
|
SRPK2
|
SRSF protein kinase 2 |
| chr19_-_45004556 | 0.13 |
ENST00000587047.1
ENST00000391956.4 ENST00000221327.4 ENST00000586637.1 ENST00000591064.1 ENST00000592529.1 |
ZNF180
|
zinc finger protein 180 |
| chr4_+_95128748 | 0.13 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr11_+_327171 | 0.13 |
ENST00000534483.1
ENST00000524824.1 ENST00000531076.1 |
RP11-326C3.12
|
RP11-326C3.12 |
| chr5_-_34919094 | 0.13 |
ENST00000341754.4
|
RAD1
|
RAD1 homolog (S. pombe) |
| chr6_+_27925019 | 0.13 |
ENST00000244623.1
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6 |
| chr7_-_32534850 | 0.13 |
ENST00000409952.3
ENST00000409909.3 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr6_-_34639733 | 0.13 |
ENST00000374021.1
|
C6orf106
|
chromosome 6 open reading frame 106 |
| chr22_+_29168652 | 0.12 |
ENST00000249064.4
ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117
|
coiled-coil domain containing 117 |
| chr12_+_133757995 | 0.12 |
ENST00000536435.2
ENST00000228289.5 ENST00000541211.2 ENST00000500625.3 ENST00000539248.2 ENST00000542711.2 ENST00000536899.2 ENST00000542986.2 ENST00000537565.1 ENST00000541975.2 |
ZNF268
|
zinc finger protein 268 |
| chr4_-_99851766 | 0.12 |
ENST00000450253.2
|
EIF4E
|
eukaryotic translation initiation factor 4E |
| chr20_+_31407692 | 0.12 |
ENST00000375571.5
|
MAPRE1
|
microtubule-associated protein, RP/EB family, member 1 |
| chr12_+_118454500 | 0.12 |
ENST00000537315.1
ENST00000229043.3 ENST00000484086.2 ENST00000420967.1 ENST00000454402.2 ENST00000392542.2 ENST00000535092.1 |
RFC5
|
replication factor C (activator 1) 5, 36.5kDa |
| chr15_-_52944231 | 0.12 |
ENST00000546305.2
|
FAM214A
|
family with sequence similarity 214, member A |
| chr12_+_133758115 | 0.12 |
ENST00000541009.2
ENST00000592241.1 |
ZNF268
|
zinc finger protein 268 |
| chr4_-_99850243 | 0.11 |
ENST00000280892.6
ENST00000511644.1 ENST00000504432.1 ENST00000505992.1 |
EIF4E
|
eukaryotic translation initiation factor 4E |
| chr3_+_150126101 | 0.11 |
ENST00000361875.3
ENST00000361136.2 |
TSC22D2
|
TSC22 domain family, member 2 |
| chr21_-_40817645 | 0.11 |
ENST00000438404.1
ENST00000358268.2 ENST00000411566.1 ENST00000451131.1 ENST00000418018.1 ENST00000415863.1 ENST00000426783.1 ENST00000288350.3 ENST00000485895.2 ENST00000448288.2 ENST00000456017.1 ENST00000434281.1 |
LCA5L
|
Leber congenital amaurosis 5-like |
| chr1_+_204494618 | 0.11 |
ENST00000367180.1
ENST00000391947.2 |
MDM4
|
Mdm4 p53 binding protein homolog (mouse) |
| chr12_-_53601000 | 0.11 |
ENST00000338737.4
ENST00000549086.2 |
ITGB7
|
integrin, beta 7 |
| chr19_-_44079611 | 0.11 |
ENST00000594107.1
ENST00000543982.1 |
XRCC1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr11_+_120255997 | 0.10 |
ENST00000532993.1
|
ARHGEF12
|
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr12_-_122107549 | 0.10 |
ENST00000355329.3
|
MORN3
|
MORN repeat containing 3 |
| chr15_+_75628419 | 0.10 |
ENST00000567377.1
ENST00000562789.1 ENST00000568301.1 |
COMMD4
|
COMM domain containing 4 |
| chr12_+_27849378 | 0.10 |
ENST00000310791.2
|
REP15
|
RAB15 effector protein |
| chr2_-_11606275 | 0.10 |
ENST00000381525.3
ENST00000362009.4 |
E2F6
|
E2F transcription factor 6 |
| chr17_-_39041479 | 0.10 |
ENST00000167588.3
|
KRT20
|
keratin 20 |
| chr2_+_242289502 | 0.10 |
ENST00000451310.1
|
SEPT2
|
septin 2 |
| chr5_-_175395283 | 0.10 |
ENST00000513482.1
ENST00000265097.4 |
THOC3
|
THO complex 3 |
| chr20_-_18477862 | 0.10 |
ENST00000337227.4
|
RBBP9
|
retinoblastoma binding protein 9 |
| chr2_+_172544011 | 0.09 |
ENST00000508530.1
|
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr4_-_185275104 | 0.09 |
ENST00000317596.3
|
RP11-290F5.2
|
RP11-290F5.2 |
| chr8_-_33370607 | 0.09 |
ENST00000360742.5
ENST00000523305.1 |
TTI2
|
TELO2 interacting protein 2 |
| chr3_-_156272924 | 0.09 |
ENST00000467789.1
ENST00000265044.2 |
SSR3
|
signal sequence receptor, gamma (translocon-associated protein gamma) |
| chr7_+_112120908 | 0.09 |
ENST00000439068.2
ENST00000312849.4 ENST00000429049.1 |
LSMEM1
|
leucine-rich single-pass membrane protein 1 |
| chr1_-_100231349 | 0.09 |
ENST00000287474.5
ENST00000414213.1 |
FRRS1
|
ferric-chelate reductase 1 |
| chr1_+_33439268 | 0.09 |
ENST00000594612.1
|
FKSG48
|
FKSG48 |
| chr7_+_7606497 | 0.09 |
ENST00000340080.4
ENST00000405785.1 ENST00000433635.1 |
MIOS
|
missing oocyte, meiosis regulator, homolog (Drosophila) |
| chr10_+_7745303 | 0.09 |
ENST00000429820.1
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr10_+_7745232 | 0.09 |
ENST00000358415.4
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr8_-_74205851 | 0.09 |
ENST00000396467.1
|
RPL7
|
ribosomal protein L7 |
| chr4_+_96012614 | 0.09 |
ENST00000264568.4
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr1_-_44818599 | 0.08 |
ENST00000537474.1
|
ERI3
|
ERI1 exoribonuclease family member 3 |
| chr6_+_37897735 | 0.08 |
ENST00000373389.5
|
ZFAND3
|
zinc finger, AN1-type domain 3 |
| chr19_+_21264980 | 0.08 |
ENST00000596053.1
ENST00000597086.1 ENST00000596143.1 ENST00000596367.1 ENST00000601416.1 |
ZNF714
|
zinc finger protein 714 |
| chr2_-_242556900 | 0.08 |
ENST00000402545.1
ENST00000402136.1 |
THAP4
|
THAP domain containing 4 |
| chr2_+_128458514 | 0.08 |
ENST00000310981.4
|
SFT2D3
|
SFT2 domain containing 3 |
| chr12_+_32832134 | 0.08 |
ENST00000452533.2
|
DNM1L
|
dynamin 1-like |
| chr19_+_42041702 | 0.08 |
ENST00000487420.2
|
AC006129.4
|
AC006129.4 |
| chr13_+_76123883 | 0.08 |
ENST00000377595.3
|
UCHL3
|
ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) |
| chr14_-_105708942 | 0.08 |
ENST00000549655.1
|
BRF1
|
BRF1, RNA polymerase III transcription initiation factor 90 kDa subunit |
| chr9_+_131901661 | 0.08 |
ENST00000423100.1
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr2_+_191334212 | 0.07 |
ENST00000444317.1
ENST00000535751.1 |
MFSD6
|
major facilitator superfamily domain containing 6 |
| chr1_+_178310581 | 0.07 |
ENST00000462775.1
|
RASAL2
|
RAS protein activator like 2 |
| chr1_+_32608566 | 0.07 |
ENST00000545542.1
|
KPNA6
|
karyopherin alpha 6 (importin alpha 7) |
| chr19_+_36239576 | 0.07 |
ENST00000587751.1
|
LIN37
|
lin-37 homolog (C. elegans) |
| chr3_-_137893721 | 0.07 |
ENST00000505015.2
ENST00000260803.4 |
DBR1
|
debranching RNA lariats 1 |
| chr16_-_70239683 | 0.07 |
ENST00000601706.1
|
AC009060.1
|
Uncharacterized protein |
| chr3_+_130569592 | 0.07 |
ENST00000533801.2
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr15_+_75628394 | 0.07 |
ENST00000564815.1
ENST00000338995.6 |
COMMD4
|
COMM domain containing 4 |
| chr2_+_172543967 | 0.07 |
ENST00000534253.2
ENST00000263811.4 ENST00000397119.3 ENST00000410079.3 ENST00000438879.1 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr11_+_67351019 | 0.07 |
ENST00000398606.3
|
GSTP1
|
glutathione S-transferase pi 1 |
| chr5_-_54988448 | 0.07 |
ENST00000503817.1
ENST00000512595.1 |
SLC38A9
|
solute carrier family 38, member 9 |
| chr3_+_51663407 | 0.07 |
ENST00000432863.1
ENST00000296477.3 |
RAD54L2
|
RAD54-like 2 (S. cerevisiae) |
| chr4_+_86699834 | 0.07 |
ENST00000395183.2
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr10_+_5005445 | 0.07 |
ENST00000380872.4
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr12_-_102224704 | 0.07 |
ENST00000299314.7
|
GNPTAB
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr6_+_88117683 | 0.07 |
ENST00000369562.4
|
C6ORF165
|
UPF0704 protein C6orf165 |
| chr19_+_46531127 | 0.06 |
ENST00000601033.1
|
CTC-344H19.4
|
CTC-344H19.4 |
| chr16_-_85722530 | 0.06 |
ENST00000253462.3
|
GINS2
|
GINS complex subunit 2 (Psf2 homolog) |
| chr3_-_93747425 | 0.06 |
ENST00000315099.2
|
STX19
|
syntaxin 19 |
| chr11_+_67351213 | 0.06 |
ENST00000398603.1
|
GSTP1
|
glutathione S-transferase pi 1 |
| chr3_+_57875738 | 0.06 |
ENST00000417128.1
ENST00000438794.1 |
SLMAP
|
sarcolemma associated protein |
| chr2_+_172543919 | 0.06 |
ENST00000452242.1
ENST00000340296.4 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr2_-_206950996 | 0.06 |
ENST00000414320.1
|
INO80D
|
INO80 complex subunit D |
| chr16_+_81070792 | 0.06 |
ENST00000564241.1
ENST00000565237.1 |
ATMIN
|
ATM interactor |
| chr9_+_131901710 | 0.06 |
ENST00000524946.2
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr7_+_134464414 | 0.06 |
ENST00000361901.2
|
CALD1
|
caldesmon 1 |
| chr17_-_295730 | 0.06 |
ENST00000329099.4
|
FAM101B
|
family with sequence similarity 101, member B |
| chr5_+_159848807 | 0.06 |
ENST00000352433.5
|
PTTG1
|
pituitary tumor-transforming 1 |
| chr12_-_11062161 | 0.06 |
ENST00000390677.2
|
TAS2R13
|
taste receptor, type 2, member 13 |
| chr21_+_40817749 | 0.06 |
ENST00000380637.3
ENST00000380634.1 ENST00000458295.1 ENST00000440288.2 ENST00000380631.1 |
SH3BGR
|
SH3 domain binding glutamic acid-rich protein |
| chr19_+_34895289 | 0.06 |
ENST00000246535.3
|
PDCD2L
|
programmed cell death 2-like |
| chr15_-_85197501 | 0.06 |
ENST00000434634.2
|
WDR73
|
WD repeat domain 73 |
| chr4_-_88244049 | 0.06 |
ENST00000328546.4
|
HSD17B13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr15_+_75628232 | 0.06 |
ENST00000267935.8
ENST00000567195.1 |
COMMD4
|
COMM domain containing 4 |
| chr2_+_42104692 | 0.06 |
ENST00000398796.2
ENST00000442214.1 |
AC104654.1
|
AC104654.1 |
| chr17_-_39341594 | 0.06 |
ENST00000398472.1
|
KRTAP4-1
|
keratin associated protein 4-1 |
| chr1_+_200011711 | 0.06 |
ENST00000544748.1
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr12_-_25150409 | 0.06 |
ENST00000549262.1
|
C12orf77
|
chromosome 12 open reading frame 77 |
| chr10_+_5005598 | 0.05 |
ENST00000442997.1
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr21_-_35284635 | 0.05 |
ENST00000429238.1
|
AP000304.12
|
AP000304.12 |
| chr5_+_159848854 | 0.05 |
ENST00000517480.1
ENST00000520452.1 ENST00000393964.1 |
PTTG1
|
pituitary tumor-transforming 1 |
| chr19_+_45690646 | 0.05 |
ENST00000591569.1
|
AC006126.3
|
Uncharacterized protein |
| chr8_+_67687413 | 0.05 |
ENST00000521960.1
ENST00000522398.1 ENST00000522629.1 ENST00000520976.1 ENST00000396596.1 |
SGK3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr3_-_138312971 | 0.05 |
ENST00000485115.1
ENST00000484888.1 ENST00000468900.1 ENST00000542237.1 ENST00000481834.1 |
CEP70
|
centrosomal protein 70kDa |
| chr17_-_18266818 | 0.05 |
ENST00000583780.1
|
SHMT1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr18_+_33709834 | 0.05 |
ENST00000358232.6
ENST00000351393.6 ENST00000442325.2 ENST00000423854.2 ENST00000350494.6 ENST00000542824.1 |
ELP2
|
elongator acetyltransferase complex subunit 2 |
| chr12_+_32832203 | 0.05 |
ENST00000553257.1
ENST00000549701.1 ENST00000358214.5 ENST00000266481.6 ENST00000551476.1 ENST00000550154.1 ENST00000547312.1 ENST00000414834.2 ENST00000381000.4 ENST00000548750.1 |
DNM1L
|
dynamin 1-like |
| chr10_+_52094298 | 0.05 |
ENST00000595931.1
|
AC069547.1
|
HCG1745369; PRO3073; Uncharacterized protein |
| chr1_-_115292591 | 0.05 |
ENST00000438362.2
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr6_+_134758827 | 0.05 |
ENST00000431422.1
|
LINC01010
|
long intergenic non-protein coding RNA 1010 |
| chr3_+_184529929 | 0.05 |
ENST00000287546.4
ENST00000437079.3 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr7_-_7606626 | 0.05 |
ENST00000609497.1
|
RP5-1159O4.1
|
RP5-1159O4.1 |
| chr6_-_165723088 | 0.05 |
ENST00000230301.8
|
C6orf118
|
chromosome 6 open reading frame 118 |
| chr10_-_75226166 | 0.05 |
ENST00000544628.1
|
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr3_+_184529948 | 0.05 |
ENST00000436792.2
ENST00000446204.2 ENST00000422105.1 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr12_-_27090896 | 0.05 |
ENST00000539625.1
ENST00000538727.1 |
ASUN
|
asunder spermatogenesis regulator |
| chr13_+_77564795 | 0.05 |
ENST00000377453.3
|
CLN5
|
ceroid-lipofuscinosis, neuronal 5 |
| chr12_+_53836339 | 0.04 |
ENST00000549135.1
|
PRR13
|
proline rich 13 |
| chr4_-_88244010 | 0.04 |
ENST00000302219.6
|
HSD17B13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr6_+_136172820 | 0.04 |
ENST00000308191.6
|
PDE7B
|
phosphodiesterase 7B |
| chr19_+_24269981 | 0.04 |
ENST00000339642.6
ENST00000357002.4 |
ZNF254
|
zinc finger protein 254 |
| chr4_-_166034029 | 0.04 |
ENST00000306480.6
|
TMEM192
|
transmembrane protein 192 |
| chr9_-_110540419 | 0.04 |
ENST00000398726.3
|
AL162389.1
|
Uncharacterized protein |
| chr10_-_74283694 | 0.04 |
ENST00000398763.4
ENST00000418483.2 ENST00000489666.2 |
MICU1
|
mitochondrial calcium uptake 1 |
| chr17_-_18266797 | 0.04 |
ENST00000316694.3
ENST00000539052.1 |
SHMT1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr11_-_111649015 | 0.04 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr15_-_55562451 | 0.04 |
ENST00000568803.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr10_+_134258649 | 0.04 |
ENST00000392630.3
ENST00000321248.2 |
C10orf91
|
chromosome 10 open reading frame 91 |
| chr14_+_58754751 | 0.04 |
ENST00000598233.1
|
AL132989.1
|
AL132989.1 |
| chr18_+_7946941 | 0.04 |
ENST00000444013.1
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chr15_-_74658519 | 0.04 |
ENST00000450547.1
ENST00000358632.4 |
CYP11A1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr15_-_74658493 | 0.03 |
ENST00000419019.2
ENST00000569662.1 |
CYP11A1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chrX_-_2882296 | 0.03 |
ENST00000438544.1
ENST00000381134.3 ENST00000545496.1 |
ARSE
|
arylsulfatase E (chondrodysplasia punctata 1) |
| chr1_+_110993795 | 0.03 |
ENST00000271331.3
|
PROK1
|
prokineticin 1 |
| chr12_-_15865844 | 0.03 |
ENST00000543612.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr4_-_39033963 | 0.03 |
ENST00000381938.3
|
TMEM156
|
transmembrane protein 156 |
| chr22_-_36236623 | 0.03 |
ENST00000405409.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr2_-_61245363 | 0.03 |
ENST00000316752.6
|
PUS10
|
pseudouridylate synthase 10 |
| chr6_+_74171301 | 0.03 |
ENST00000415954.2
ENST00000498286.1 ENST00000370305.1 ENST00000370300.4 |
MTO1
|
mitochondrial tRNA translation optimization 1 |
| chr6_+_42749759 | 0.03 |
ENST00000314073.5
|
GLTSCR1L
|
GLTSCR1-like |
| chr5_+_35856951 | 0.03 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr12_+_21207503 | 0.03 |
ENST00000545916.1
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr1_+_100731819 | 0.03 |
ENST00000370126.1
|
RTCA
|
RNA 3'-terminal phosphate cyclase |
| chr3_-_138313161 | 0.03 |
ENST00000489254.1
ENST00000474781.1 ENST00000462419.1 ENST00000264982.3 |
CEP70
|
centrosomal protein 70kDa |
| chr12_-_53601055 | 0.03 |
ENST00000552972.1
ENST00000422257.3 ENST00000267082.5 |
ITGB7
|
integrin, beta 7 |
| chr9_+_79074068 | 0.03 |
ENST00000444201.2
ENST00000376730.4 |
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2 |
| chr2_-_188419078 | 0.03 |
ENST00000437725.1
ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr3_-_155524049 | 0.02 |
ENST00000534941.1
ENST00000340171.2 |
C3orf33
|
chromosome 3 open reading frame 33 |
| chr17_+_40925454 | 0.02 |
ENST00000253794.2
ENST00000590339.1 ENST00000589520.1 |
VPS25
|
vacuolar protein sorting 25 homolog (S. cerevisiae) |
| chr5_+_140560980 | 0.02 |
ENST00000361016.2
|
PCDHB16
|
protocadherin beta 16 |
| chr16_+_68071816 | 0.02 |
ENST00000562246.1
|
DUS2
|
dihydrouridine synthase 2 |
| chr5_+_140705777 | 0.02 |
ENST00000606901.1
ENST00000606674.1 |
AC005618.6
|
AC005618.6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB16
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0046022 | positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.1 | 0.5 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.1 | 0.4 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.2 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 0.2 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.2 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.1 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.0 | 0.3 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.0 | 0.1 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.3 | GO:1903899 | lung goblet cell differentiation(GO:0060480) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.1 | GO:1904875 | regulation of DNA ligase activity(GO:1904875) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.1 | GO:0051598 | meiotic recombination checkpoint(GO:0051598) |
| 0.0 | 0.2 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.2 | GO:0015853 | adenine transport(GO:0015853) |
| 0.0 | 0.1 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.4 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.3 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.1 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.0 | 0.1 | GO:1904482 | response to tetrahydrofolate(GO:1904481) cellular response to tetrahydrofolate(GO:1904482) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.0 | 0.2 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.1 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.2 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.1 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.2 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.2 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0015322 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.1 | 0.3 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 0.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.2 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.2 | GO:0033265 | acetylcholinesterase activity(GO:0003990) choline binding(GO:0033265) |
| 0.0 | 0.2 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.2 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.0 | 0.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.1 | GO:0070026 | nitric oxide binding(GO:0070026) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.5 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.0 | 0.1 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.0 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |