Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
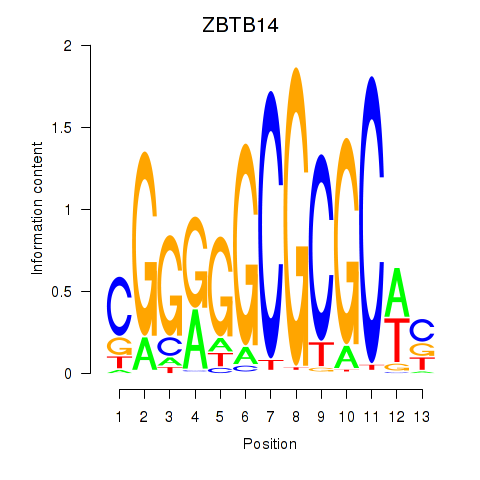
Results for ZBTB14
Z-value: 0.81
Transcription factors associated with ZBTB14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB14
|
ENSG00000198081.6 | zinc finger and BTB domain containing 14 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB14 | hg19_v2_chr18_-_5296138_5296194 | -0.22 | 7.8e-01 | Click! |
Activity profile of ZBTB14 motif
Sorted Z-values of ZBTB14 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_78740511 | 0.72 |
ENST00000504123.1
ENST00000264903.4 ENST00000515441.1 |
CNOT6L
|
CCR4-NOT transcription complex, subunit 6-like |
| chr10_-_977564 | 0.44 |
ENST00000406525.2
|
LARP4B
|
La ribonucleoprotein domain family, member 4B |
| chr4_+_4861385 | 0.36 |
ENST00000382723.4
|
MSX1
|
msh homeobox 1 |
| chr20_-_56284816 | 0.36 |
ENST00000395819.3
ENST00000341744.3 |
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr12_+_112451222 | 0.35 |
ENST00000552052.1
|
ERP29
|
endoplasmic reticulum protein 29 |
| chr14_+_91526668 | 0.35 |
ENST00000521334.1
|
C14orf159
|
chromosome 14 open reading frame 159 |
| chr8_+_25042192 | 0.35 |
ENST00000410074.1
|
DOCK5
|
dedicator of cytokinesis 5 |
| chr1_-_223537475 | 0.32 |
ENST00000344029.6
ENST00000494793.2 ENST00000366878.4 ENST00000366877.3 |
SUSD4
|
sushi domain containing 4 |
| chr11_-_33891362 | 0.29 |
ENST00000395833.3
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr5_-_134914673 | 0.29 |
ENST00000512158.1
|
CXCL14
|
chemokine (C-X-C motif) ligand 14 |
| chr7_+_156931889 | 0.28 |
ENST00000389103.4
|
UBE3C
|
ubiquitin protein ligase E3C |
| chr3_-_107809816 | 0.27 |
ENST00000361309.5
ENST00000355354.7 |
CD47
|
CD47 molecule |
| chr21_-_46237883 | 0.26 |
ENST00000397893.3
|
SUMO3
|
small ubiquitin-like modifier 3 |
| chr1_+_178694300 | 0.25 |
ENST00000367635.3
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr17_-_38520067 | 0.25 |
ENST00000337376.4
ENST00000578689.1 |
GJD3
|
gap junction protein, delta 3, 31.9kDa |
| chr6_-_83775442 | 0.24 |
ENST00000369745.5
|
UBE3D
|
ubiquitin protein ligase E3D |
| chr17_-_42277203 | 0.24 |
ENST00000587097.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr17_-_66287310 | 0.23 |
ENST00000582867.1
|
SLC16A6
|
solute carrier family 16, member 6 |
| chr3_-_133614597 | 0.22 |
ENST00000285208.4
ENST00000460865.3 |
RAB6B
|
RAB6B, member RAS oncogene family |
| chr9_-_38069208 | 0.22 |
ENST00000377707.3
ENST00000377700.4 |
SHB
|
Src homology 2 domain containing adaptor protein B |
| chr11_-_47788985 | 0.22 |
ENST00000540172.2
|
FNBP4
|
formin binding protein 4 |
| chr5_+_98264867 | 0.22 |
ENST00000513175.1
|
CTD-2007H13.3
|
CTD-2007H13.3 |
| chr3_-_138048653 | 0.22 |
ENST00000460099.1
|
NME9
|
NME/NM23 family member 9 |
| chr13_-_52027134 | 0.21 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr12_+_133067157 | 0.21 |
ENST00000261673.6
|
FBRSL1
|
fibrosin-like 1 |
| chr6_-_38607673 | 0.21 |
ENST00000481247.1
|
BTBD9
|
BTB (POZ) domain containing 9 |
| chr1_+_6845384 | 0.21 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr12_-_129308041 | 0.21 |
ENST00000376740.4
|
SLC15A4
|
solute carrier family 15 (oligopeptide transporter), member 4 |
| chr18_+_9102592 | 0.20 |
ENST00000318388.6
|
NDUFV2
|
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
| chr15_-_59041954 | 0.20 |
ENST00000439637.1
ENST00000558004.1 |
ADAM10
|
ADAM metallopeptidase domain 10 |
| chr16_+_56623433 | 0.19 |
ENST00000570176.1
|
MT3
|
metallothionein 3 |
| chr3_+_184016986 | 0.19 |
ENST00000417952.1
|
PSMD2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| chr14_-_91526922 | 0.19 |
ENST00000418736.2
ENST00000261991.3 |
RPS6KA5
|
ribosomal protein S6 kinase, 90kDa, polypeptide 5 |
| chr20_+_49348109 | 0.19 |
ENST00000396039.1
|
PARD6B
|
par-6 family cell polarity regulator beta |
| chr18_-_51750948 | 0.18 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr9_-_115095851 | 0.18 |
ENST00000343327.2
|
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr2_-_203103185 | 0.18 |
ENST00000409205.1
|
SUMO1
|
small ubiquitin-like modifier 1 |
| chr4_+_56212505 | 0.17 |
ENST00000505210.1
|
SRD5A3
|
steroid 5 alpha-reductase 3 |
| chr7_-_45960850 | 0.17 |
ENST00000381083.4
ENST00000381086.5 ENST00000275521.6 |
IGFBP3
|
insulin-like growth factor binding protein 3 |
| chr18_+_9102669 | 0.17 |
ENST00000497577.2
|
NDUFV2
|
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
| chr12_+_111843749 | 0.17 |
ENST00000341259.2
|
SH2B3
|
SH2B adaptor protein 3 |
| chr16_+_30709530 | 0.16 |
ENST00000411466.2
|
SRCAP
|
Snf2-related CREBBP activator protein |
| chr8_+_25042268 | 0.16 |
ENST00000481100.1
ENST00000276440.7 |
DOCK5
|
dedicator of cytokinesis 5 |
| chr2_+_232573208 | 0.16 |
ENST00000409115.3
|
PTMA
|
prothymosin, alpha |
| chr19_+_35521616 | 0.16 |
ENST00000595652.1
|
SCN1B
|
sodium channel, voltage-gated, type I, beta subunit |
| chr15_-_71146460 | 0.16 |
ENST00000344870.4
|
LARP6
|
La ribonucleoprotein domain family, member 6 |
| chr3_+_194406603 | 0.16 |
ENST00000329759.4
|
FAM43A
|
family with sequence similarity 43, member A |
| chr2_-_17935027 | 0.16 |
ENST00000446852.1
|
SMC6
|
structural maintenance of chromosomes 6 |
| chr2_-_128145498 | 0.16 |
ENST00000409179.2
|
MAP3K2
|
mitogen-activated protein kinase kinase kinase 2 |
| chr22_+_46972975 | 0.16 |
ENST00000431155.1
|
GRAMD4
|
GRAM domain containing 4 |
| chr1_-_33336414 | 0.15 |
ENST00000373471.3
ENST00000609187.1 |
FNDC5
|
fibronectin type III domain containing 5 |
| chr1_-_223537401 | 0.15 |
ENST00000343846.3
ENST00000454695.2 ENST00000484758.2 |
SUSD4
|
sushi domain containing 4 |
| chrX_+_11776410 | 0.15 |
ENST00000361672.2
|
MSL3
|
male-specific lethal 3 homolog (Drosophila) |
| chr4_+_17578815 | 0.15 |
ENST00000226299.4
|
LAP3
|
leucine aminopeptidase 3 |
| chr16_-_89268070 | 0.15 |
ENST00000562855.2
|
SLC22A31
|
solute carrier family 22, member 31 |
| chr3_+_14444063 | 0.15 |
ENST00000454876.2
ENST00000360861.3 ENST00000416216.2 |
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter), member 6 |
| chr18_-_3013307 | 0.15 |
ENST00000584294.1
|
LPIN2
|
lipin 2 |
| chr3_+_23244511 | 0.15 |
ENST00000425792.1
|
UBE2E2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr4_-_54930790 | 0.15 |
ENST00000263921.3
|
CHIC2
|
cysteine-rich hydrophobic domain 2 |
| chr18_+_9913977 | 0.15 |
ENST00000400000.2
ENST00000340541.4 |
VAPA
|
VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa |
| chr2_+_232573222 | 0.14 |
ENST00000341369.7
ENST00000409683.1 |
PTMA
|
prothymosin, alpha |
| chr19_+_1286097 | 0.14 |
ENST00000215368.2
|
EFNA2
|
ephrin-A2 |
| chr11_-_61584233 | 0.14 |
ENST00000491310.1
|
FADS1
|
fatty acid desaturase 1 |
| chr1_+_6845497 | 0.14 |
ENST00000473578.1
ENST00000557126.1 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr19_+_35521572 | 0.14 |
ENST00000262631.5
|
SCN1B
|
sodium channel, voltage-gated, type I, beta subunit |
| chr16_+_66461175 | 0.14 |
ENST00000536005.2
ENST00000299694.8 ENST00000561796.1 |
BEAN1
|
brain expressed, associated with NEDD4, 1 |
| chr9_+_137218362 | 0.14 |
ENST00000481739.1
|
RXRA
|
retinoid X receptor, alpha |
| chr18_-_12884150 | 0.14 |
ENST00000591115.1
ENST00000309660.5 |
PTPN2
|
protein tyrosine phosphatase, non-receptor type 2 |
| chr19_+_35521699 | 0.14 |
ENST00000415950.3
|
SCN1B
|
sodium channel, voltage-gated, type I, beta subunit |
| chr5_+_65440032 | 0.14 |
ENST00000334121.6
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr17_-_31204124 | 0.14 |
ENST00000579584.1
ENST00000318217.5 ENST00000583621.1 |
MYO1D
|
myosin ID |
| chr9_+_88556444 | 0.14 |
ENST00000376040.1
|
NAA35
|
N(alpha)-acetyltransferase 35, NatC auxiliary subunit |
| chr17_-_66287257 | 0.14 |
ENST00000327268.4
|
SLC16A6
|
solute carrier family 16, member 6 |
| chr1_+_200842083 | 0.14 |
ENST00000304244.2
|
GPR25
|
G protein-coupled receptor 25 |
| chr2_+_232575168 | 0.13 |
ENST00000440384.1
|
PTMA
|
prothymosin, alpha |
| chrX_-_71525742 | 0.13 |
ENST00000450875.1
ENST00000417400.1 ENST00000431381.1 ENST00000445983.1 |
CITED1
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chr9_+_6758109 | 0.13 |
ENST00000536108.1
|
KDM4C
|
lysine (K)-specific demethylase 4C |
| chr18_+_158513 | 0.13 |
ENST00000400266.3
ENST00000580410.1 ENST00000383589.2 ENST00000261601.7 |
USP14
|
ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase) |
| chr19_+_18529674 | 0.13 |
ENST00000597724.2
|
SSBP4
|
single stranded DNA binding protein 4 |
| chr3_+_170075436 | 0.13 |
ENST00000476188.1
ENST00000259119.4 ENST00000426052.2 |
SKIL
|
SKI-like oncogene |
| chr11_-_129149197 | 0.13 |
ENST00000525234.1
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr15_-_71146347 | 0.13 |
ENST00000559140.2
|
LARP6
|
La ribonucleoprotein domain family, member 6 |
| chr5_+_176560595 | 0.13 |
ENST00000508896.1
|
NSD1
|
nuclear receptor binding SET domain protein 1 |
| chr14_+_102027688 | 0.13 |
ENST00000510508.4
ENST00000359323.3 |
DIO3
|
deiodinase, iodothyronine, type III |
| chr7_+_65338312 | 0.13 |
ENST00000434382.2
|
VKORC1L1
|
vitamin K epoxide reductase complex, subunit 1-like 1 |
| chr9_+_139606983 | 0.13 |
ENST00000371692.4
|
FAM69B
|
family with sequence similarity 69, member B |
| chr19_-_54693146 | 0.12 |
ENST00000414665.1
ENST00000453320.1 |
MBOAT7
|
membrane bound O-acyltransferase domain containing 7 |
| chr16_-_75285762 | 0.12 |
ENST00000564028.1
|
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr8_-_23315131 | 0.12 |
ENST00000518718.1
|
ENTPD4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr11_-_10879593 | 0.12 |
ENST00000528289.1
ENST00000432999.2 |
ZBED5
|
zinc finger, BED-type containing 5 |
| chr3_+_23244579 | 0.12 |
ENST00000452894.1
|
UBE2E2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr3_+_170136642 | 0.12 |
ENST00000064724.3
ENST00000486975.1 |
CLDN11
|
claudin 11 |
| chr4_-_15656923 | 0.12 |
ENST00000382358.4
ENST00000412094.2 ENST00000341285.3 |
FBXL5
|
F-box and leucine-rich repeat protein 5 |
| chr2_-_153573887 | 0.12 |
ENST00000493468.2
ENST00000545856.1 |
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr4_-_119757322 | 0.12 |
ENST00000379735.5
|
SEC24D
|
SEC24 family member D |
| chr12_-_112450915 | 0.12 |
ENST00000437003.2
ENST00000552374.2 ENST00000550831.3 ENST00000354825.3 ENST00000549537.2 ENST00000355445.3 |
TMEM116
|
transmembrane protein 116 |
| chr9_+_8858102 | 0.12 |
ENST00000447950.1
ENST00000430766.1 |
RP11-75C9.1
|
RP11-75C9.1 |
| chr2_+_28113583 | 0.12 |
ENST00000344773.2
ENST00000379624.1 ENST00000342045.2 ENST00000379632.2 ENST00000361704.2 |
BRE
|
brain and reproductive organ-expressed (TNFRSF1A modulator) |
| chr12_+_13197218 | 0.12 |
ENST00000197268.8
|
KIAA1467
|
KIAA1467 |
| chr1_+_182992545 | 0.12 |
ENST00000258341.4
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2) |
| chr1_+_174128536 | 0.12 |
ENST00000357444.6
ENST00000367689.3 |
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr3_-_194207388 | 0.12 |
ENST00000457986.1
|
ATP13A3
|
ATPase type 13A3 |
| chr2_+_74741569 | 0.12 |
ENST00000233638.7
|
TLX2
|
T-cell leukemia homeobox 2 |
| chr7_-_55640141 | 0.12 |
ENST00000452832.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chrX_+_135579670 | 0.12 |
ENST00000218364.4
|
HTATSF1
|
HIV-1 Tat specific factor 1 |
| chr12_+_64798095 | 0.12 |
ENST00000332707.5
|
XPOT
|
exportin, tRNA |
| chr9_+_103189458 | 0.12 |
ENST00000398977.2
|
MSANTD3
|
Myb/SANT-like DNA-binding domain containing 3 |
| chr22_+_19744226 | 0.12 |
ENST00000332710.4
ENST00000329705.7 ENST00000359500.3 |
TBX1
|
T-box 1 |
| chr5_+_61602236 | 0.12 |
ENST00000514082.1
ENST00000407818.3 |
KIF2A
|
kinesin heavy chain member 2A |
| chr18_-_12884259 | 0.12 |
ENST00000353319.4
ENST00000327283.3 |
PTPN2
|
protein tyrosine phosphatase, non-receptor type 2 |
| chr2_+_25264933 | 0.12 |
ENST00000401432.3
ENST00000403714.3 |
EFR3B
|
EFR3 homolog B (S. cerevisiae) |
| chr1_-_91487806 | 0.12 |
ENST00000361321.5
|
ZNF644
|
zinc finger protein 644 |
| chr15_-_71407833 | 0.11 |
ENST00000449977.2
|
CT62
|
cancer/testis antigen 62 |
| chr7_-_139876734 | 0.11 |
ENST00000006967.5
|
JHDM1D
|
lysine (K)-specific demethylase 7A |
| chr8_-_90996837 | 0.11 |
ENST00000519426.1
ENST00000265433.3 |
NBN
|
nibrin |
| chrX_+_70364667 | 0.11 |
ENST00000536169.1
ENST00000395855.2 ENST00000374051.3 ENST00000358741.3 |
NLGN3
|
neuroligin 3 |
| chr6_-_19804973 | 0.11 |
ENST00000457670.1
ENST00000607810.1 ENST00000606628.1 |
RP4-625H18.2
|
RP4-625H18.2 |
| chr19_-_19006890 | 0.11 |
ENST00000247005.6
|
GDF1
|
growth differentiation factor 1 |
| chr12_+_95611536 | 0.11 |
ENST00000549002.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr16_+_54964740 | 0.11 |
ENST00000394636.4
|
IRX5
|
iroquois homeobox 5 |
| chr11_-_64512273 | 0.11 |
ENST00000377497.3
ENST00000377487.1 ENST00000430645.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr1_+_185014647 | 0.11 |
ENST00000367509.4
|
RNF2
|
ring finger protein 2 |
| chr7_+_77166592 | 0.11 |
ENST00000248594.6
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr19_+_1941117 | 0.11 |
ENST00000255641.8
|
CSNK1G2
|
casein kinase 1, gamma 2 |
| chr1_-_111682813 | 0.11 |
ENST00000539140.1
|
DRAM2
|
DNA-damage regulated autophagy modulator 2 |
| chr8_+_12809531 | 0.11 |
ENST00000532376.2
|
KIAA1456
|
KIAA1456 |
| chr4_-_119757239 | 0.11 |
ENST00000280551.6
|
SEC24D
|
SEC24 family member D |
| chr9_-_4299874 | 0.11 |
ENST00000381971.3
ENST00000477901.1 |
GLIS3
|
GLIS family zinc finger 3 |
| chr5_+_179125907 | 0.11 |
ENST00000247461.4
ENST00000452673.2 ENST00000502498.1 ENST00000507307.1 ENST00000513246.1 ENST00000502673.1 ENST00000506654.1 ENST00000512607.2 ENST00000510810.1 |
CANX
|
calnexin |
| chr1_+_23037323 | 0.11 |
ENST00000544305.1
ENST00000374630.3 ENST00000400191.3 ENST00000374632.3 |
EPHB2
|
EPH receptor B2 |
| chr14_+_89029323 | 0.11 |
ENST00000554602.1
|
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr15_-_59041768 | 0.11 |
ENST00000402627.1
ENST00000396140.2 ENST00000559053.1 ENST00000561288.1 |
ADAM10
|
ADAM metallopeptidase domain 10 |
| chr20_+_9049742 | 0.11 |
ENST00000437503.1
|
PLCB4
|
phospholipase C, beta 4 |
| chr5_+_176873446 | 0.11 |
ENST00000507881.1
|
PRR7
|
proline rich 7 (synaptic) |
| chr3_+_98451093 | 0.11 |
ENST00000483910.1
ENST00000460774.1 |
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr2_+_5832799 | 0.11 |
ENST00000322002.3
|
SOX11
|
SRY (sex determining region Y)-box 11 |
| chr19_+_35634146 | 0.11 |
ENST00000586063.1
ENST00000270310.2 ENST00000588265.1 |
FXYD7
|
FXYD domain containing ion transport regulator 7 |
| chr2_+_109745655 | 0.11 |
ENST00000418513.1
|
SH3RF3
|
SH3 domain containing ring finger 3 |
| chr15_-_83735889 | 0.11 |
ENST00000379403.2
|
BTBD1
|
BTB (POZ) domain containing 1 |
| chr19_+_18530184 | 0.11 |
ENST00000601357.2
|
SSBP4
|
single stranded DNA binding protein 4 |
| chr1_+_16174280 | 0.11 |
ENST00000375759.3
|
SPEN
|
spen family transcriptional repressor |
| chr3_-_185826286 | 0.11 |
ENST00000537818.1
ENST00000422039.1 ENST00000434744.1 |
ETV5
|
ets variant 5 |
| chrX_-_47004437 | 0.10 |
ENST00000276062.8
|
NDUFB11
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11, 17.3kDa |
| chr17_+_72428266 | 0.10 |
ENST00000582473.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr20_+_46130671 | 0.10 |
ENST00000371998.3
ENST00000371997.3 |
NCOA3
|
nuclear receptor coactivator 3 |
| chr5_+_148521046 | 0.10 |
ENST00000326685.7
ENST00000356541.3 ENST00000309868.7 |
ABLIM3
|
actin binding LIM protein family, member 3 |
| chr5_+_52285144 | 0.10 |
ENST00000296585.5
|
ITGA2
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) |
| chr7_+_77167376 | 0.10 |
ENST00000435495.2
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr1_+_6845578 | 0.10 |
ENST00000467404.2
ENST00000439411.2 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr22_+_39745930 | 0.10 |
ENST00000318801.4
ENST00000216155.7 ENST00000406293.3 ENST00000328933.5 |
SYNGR1
|
synaptogyrin 1 |
| chr5_+_118604385 | 0.10 |
ENST00000274456.6
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr11_+_69455855 | 0.10 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr5_+_176873789 | 0.10 |
ENST00000323249.3
ENST00000502922.1 |
PRR7
|
proline rich 7 (synaptic) |
| chr8_+_132916318 | 0.10 |
ENST00000254624.5
ENST00000522709.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chr2_-_55646957 | 0.10 |
ENST00000263630.8
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr7_-_139876812 | 0.10 |
ENST00000397560.2
|
JHDM1D
|
lysine (K)-specific demethylase 7A |
| chr18_-_21242833 | 0.10 |
ENST00000586087.1
ENST00000592179.1 |
ANKRD29
|
ankyrin repeat domain 29 |
| chr19_+_41107249 | 0.10 |
ENST00000396819.3
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr8_-_142011316 | 0.10 |
ENST00000522684.1
ENST00000395218.2 ENST00000524357.1 ENST00000521332.1 ENST00000524040.1 ENST00000519881.1 ENST00000520045.1 |
PTK2
|
protein tyrosine kinase 2 |
| chrX_-_153200513 | 0.10 |
ENST00000432089.1
|
NAA10
|
N(alpha)-acetyltransferase 10, NatA catalytic subunit |
| chr17_+_30264014 | 0.10 |
ENST00000322652.5
ENST00000580398.1 |
SUZ12
|
SUZ12 polycomb repressive complex 2 subunit |
| chr18_+_12948000 | 0.10 |
ENST00000585730.1
ENST00000399892.2 ENST00000589446.1 ENST00000587761.1 |
SEH1L
|
SEH1-like (S. cerevisiae) |
| chr6_+_30614886 | 0.10 |
ENST00000376471.4
|
C6orf136
|
chromosome 6 open reading frame 136 |
| chr20_+_49348081 | 0.10 |
ENST00000371610.2
|
PARD6B
|
par-6 family cell polarity regulator beta |
| chr2_-_230786619 | 0.10 |
ENST00000389045.3
ENST00000409677.1 |
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr2_+_227700652 | 0.10 |
ENST00000341329.3
ENST00000392062.2 ENST00000437454.1 ENST00000443477.1 ENST00000423616.1 ENST00000448992.1 |
RHBDD1
|
rhomboid domain containing 1 |
| chr14_+_105266933 | 0.10 |
ENST00000555360.1
|
ZBTB42
|
zinc finger and BTB domain containing 42 |
| chrX_+_135229559 | 0.10 |
ENST00000394155.2
|
FHL1
|
four and a half LIM domains 1 |
| chr22_-_45559642 | 0.10 |
ENST00000426282.2
|
CTA-217C2.1
|
CTA-217C2.1 |
| chr11_+_66384053 | 0.10 |
ENST00000310137.4
ENST00000393979.3 ENST00000409372.1 ENST00000443702.1 ENST00000409738.4 ENST00000514361.3 ENST00000503028.2 ENST00000412278.2 ENST00000500635.2 |
RBM14
RBM4
RBM14-RBM4
|
RNA binding motif protein 14 RNA binding motif protein 4 RBM14-RBM4 readthrough |
| chr12_+_121078355 | 0.10 |
ENST00000316803.3
|
CABP1
|
calcium binding protein 1 |
| chrX_+_135229600 | 0.10 |
ENST00000370690.3
|
FHL1
|
four and a half LIM domains 1 |
| chr17_-_78450398 | 0.10 |
ENST00000306773.4
|
NPTX1
|
neuronal pentraxin I |
| chr1_-_78149041 | 0.10 |
ENST00000414381.1
ENST00000370798.1 |
ZZZ3
|
zinc finger, ZZ-type containing 3 |
| chr9_+_112542572 | 0.10 |
ENST00000374530.3
|
PALM2-AKAP2
|
PALM2-AKAP2 readthrough |
| chr17_+_79651007 | 0.10 |
ENST00000572392.1
ENST00000577012.1 |
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr9_+_6758024 | 0.10 |
ENST00000442236.2
|
KDM4C
|
lysine (K)-specific demethylase 4C |
| chr19_+_18530146 | 0.10 |
ENST00000348495.6
ENST00000270061.7 |
SSBP4
|
single stranded DNA binding protein 4 |
| chr11_+_57435219 | 0.09 |
ENST00000527985.1
ENST00000287169.3 |
ZDHHC5
|
zinc finger, DHHC-type containing 5 |
| chr3_-_118753792 | 0.09 |
ENST00000480431.1
|
IGSF11
|
immunoglobulin superfamily, member 11 |
| chrX_-_47004878 | 0.09 |
ENST00000377811.3
|
NDUFB11
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11, 17.3kDa |
| chr17_+_46985823 | 0.09 |
ENST00000508468.2
|
UBE2Z
|
ubiquitin-conjugating enzyme E2Z |
| chr14_-_105635090 | 0.09 |
ENST00000331782.3
ENST00000347004.2 |
JAG2
|
jagged 2 |
| chr11_+_57435441 | 0.09 |
ENST00000528177.1
|
ZDHHC5
|
zinc finger, DHHC-type containing 5 |
| chr13_+_114238997 | 0.09 |
ENST00000538138.1
ENST00000375370.5 |
TFDP1
|
transcription factor Dp-1 |
| chr3_+_98451532 | 0.09 |
ENST00000486334.2
ENST00000394162.1 |
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr8_-_23540402 | 0.09 |
ENST00000523261.1
ENST00000380871.4 |
NKX3-1
|
NK3 homeobox 1 |
| chr12_+_52345448 | 0.09 |
ENST00000257963.4
ENST00000541224.1 ENST00000426655.2 ENST00000536420.1 ENST00000415850.2 |
ACVR1B
|
activin A receptor, type IB |
| chr12_-_125052010 | 0.09 |
ENST00000458234.1
|
NCOR2
|
nuclear receptor corepressor 2 |
| chr2_+_220306745 | 0.09 |
ENST00000431523.1
ENST00000396698.1 ENST00000396695.2 |
SPEG
|
SPEG complex locus |
| chr6_-_143266297 | 0.09 |
ENST00000367603.2
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2 |
| chr19_+_42817527 | 0.09 |
ENST00000598766.1
|
TMEM145
|
transmembrane protein 145 |
| chr17_-_46703826 | 0.09 |
ENST00000550387.1
ENST00000311177.5 |
HOXB9
|
homeobox B9 |
| chr8_-_90996459 | 0.09 |
ENST00000517337.1
ENST00000409330.1 |
NBN
|
nibrin |
| chr11_-_46142615 | 0.09 |
ENST00000529734.1
ENST00000323180.6 |
PHF21A
|
PHD finger protein 21A |
| chr1_+_60280458 | 0.09 |
ENST00000455990.1
ENST00000371208.3 |
HOOK1
|
hook microtubule-tethering protein 1 |
| chr21_+_38071430 | 0.09 |
ENST00000290399.6
|
SIM2
|
single-minded family bHLH transcription factor 2 |
| chr1_+_224370873 | 0.09 |
ENST00000323699.4
ENST00000391877.3 |
DEGS1
|
delta(4)-desaturase, sphingolipid 1 |
| chr13_+_52158610 | 0.09 |
ENST00000298125.5
|
WDFY2
|
WD repeat and FYVE domain containing 2 |
| chr5_-_115177247 | 0.09 |
ENST00000500945.2
|
ATG12
|
autophagy related 12 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB14
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.1 | 0.4 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.1 | 0.3 | GO:1902232 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) regulation of positive thymic T cell selection(GO:1902232) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.1 | 0.7 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 0.2 | GO:2000374 | cadmium ion homeostasis(GO:0055073) negative regulation of hydrogen peroxide catabolic process(GO:2000296) regulation of oxygen metabolic process(GO:2000374) |
| 0.1 | 0.5 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.3 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.1 | 0.3 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.2 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.1 | GO:0021644 | vagus nerve morphogenesis(GO:0021644) |
| 0.0 | 0.3 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:0045994 | positive regulation of translational initiation by iron(GO:0045994) |
| 0.0 | 0.2 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.2 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.3 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.0 | 0.3 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.3 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.1 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.0 | 0.2 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.1 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.1 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.0 | 0.2 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.4 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.2 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 0.1 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.0 | 0.1 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.1 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.0 | 0.2 | GO:2000834 | androgen secretion(GO:0035935) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) |
| 0.0 | 0.1 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.1 | GO:0042391 | regulation of membrane potential(GO:0042391) |
| 0.0 | 0.2 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.0 | 0.1 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.0 | 0.2 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.4 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0015728 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.0 | 0.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.2 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.1 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.2 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.0 | GO:0070101 | positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.0 | 0.0 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.0 | 0.1 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.1 | GO:1901254 | modulation by host of viral RNA genome replication(GO:0044830) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.0 | 0.1 | GO:1903216 | negative regulation of late endosome to lysosome transport(GO:1902823) regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.0 | 0.4 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.1 | GO:1904717 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.1 | GO:0007068 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.0 | 0.1 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.1 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.1 | GO:2000657 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.0 | 0.1 | GO:0045081 | negative regulation of interleukin-10 biosynthetic process(GO:0045081) |
| 0.0 | 0.1 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.1 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.2 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.1 | GO:0030920 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.1 | GO:0071105 | response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.0 | 0.1 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.1 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.0 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.1 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.0 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.0 | 0.1 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 0.0 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) negative regulation of vascular smooth muscle cell differentiation(GO:1905064) |
| 0.0 | 0.0 | GO:0097065 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 0.1 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.1 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.2 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 | 0.1 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.0 | GO:0032912 | negative regulation of transforming growth factor beta2 production(GO:0032912) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0051708 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.2 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.2 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.3 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.1 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.1 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.0 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.0 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.3 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.0 | GO:1901214 | regulation of neuron death(GO:1901214) |
| 0.0 | 0.3 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.0 | GO:0016115 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.0 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.0 | GO:1900239 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.0 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 | 0.0 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.0 | GO:0045629 | positive regulation of T-helper 1 cell differentiation(GO:0045627) negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.0 | GO:0003104 | positive regulation of glomerular filtration(GO:0003104) regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.0 | 0.0 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.0 | 0.1 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.1 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.1 | GO:1904386 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.0 | GO:0021836 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.0 | GO:0001889 | liver development(GO:0001889) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.1 | 0.2 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.1 | 0.4 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.1 | 0.2 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.0 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.2 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.3 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) laminin-11 complex(GO:0043260) |
| 0.0 | 0.7 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.0 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.0 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.1 | GO:0097422 | tubular endosome(GO:0097422) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.1 | 0.2 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.1 | 0.3 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.1 | 0.2 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.1 | 0.4 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.0 | 0.2 | GO:0033765 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0045485 | omega-6 fatty acid desaturase activity(GO:0045485) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0016705 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen(GO:0016705) |
| 0.0 | 0.2 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.4 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.2 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.1 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.0 | 0.2 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.2 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.3 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.0 | 0.3 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.2 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.2 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.2 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.1 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.0 | 0.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.1 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.0 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.2 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.0 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.0 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.0 | 0.0 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0010736 | serum response element binding(GO:0010736) |
| 0.0 | 0.6 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.0 | GO:0047661 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.0 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.5 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |