Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
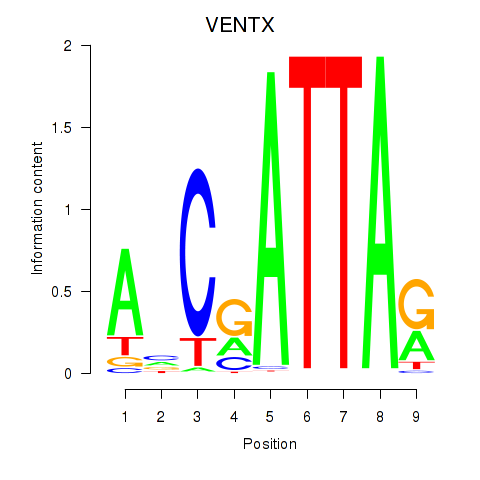
Results for VENTX
Z-value: 1.51
Transcription factors associated with VENTX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
VENTX
|
ENSG00000151650.7 | VENT homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| VENTX | hg19_v2_chr10_+_135050908_135050908 | 0.89 | 1.1e-01 | Click! |
Activity profile of VENTX motif
Sorted Z-values of VENTX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_67968793 | 1.86 |
ENST00000417488.1
|
RP11-195B21.3
|
Protein LOC644249 |
| chr4_-_155511887 | 1.70 |
ENST00000302053.3
ENST00000403106.3 |
FGA
|
fibrinogen alpha chain |
| chr8_+_107738240 | 1.36 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr10_-_33405600 | 1.26 |
ENST00000414308.1
|
RP11-342D11.3
|
RP11-342D11.3 |
| chr13_+_110958124 | 1.23 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr13_-_67802549 | 1.09 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr6_+_43968306 | 1.04 |
ENST00000442114.2
ENST00000336600.5 ENST00000439969.2 |
C6orf223
|
chromosome 6 open reading frame 223 |
| chr5_-_79950775 | 1.04 |
ENST00000439211.2
|
DHFR
|
dihydrofolate reductase |
| chr4_+_119200215 | 1.01 |
ENST00000602573.1
|
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr18_+_29171689 | 0.96 |
ENST00000237014.3
|
TTR
|
transthyretin |
| chr5_+_95066823 | 0.89 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chrX_+_106163626 | 0.88 |
ENST00000336803.1
|
CLDN2
|
claudin 2 |
| chr17_-_47045949 | 0.74 |
ENST00000357424.2
|
GIP
|
gastric inhibitory polypeptide |
| chr3_-_57233966 | 0.72 |
ENST00000473921.1
ENST00000295934.3 |
HESX1
|
HESX homeobox 1 |
| chr4_+_169418195 | 0.70 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr12_+_56473939 | 0.69 |
ENST00000450146.2
|
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr12_+_56473910 | 0.69 |
ENST00000411731.2
|
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr1_+_78470530 | 0.66 |
ENST00000370763.5
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chr4_-_105416039 | 0.66 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr9_+_2159850 | 0.65 |
ENST00000416751.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr1_+_62417957 | 0.64 |
ENST00000307297.7
ENST00000543708.1 |
INADL
|
InaD-like (Drosophila) |
| chr17_+_68100989 | 0.61 |
ENST00000585558.1
ENST00000392670.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr6_-_111927449 | 0.61 |
ENST00000368761.5
ENST00000392556.4 ENST00000340026.6 |
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr7_+_139025875 | 0.60 |
ENST00000297534.6
|
C7orf55
|
chromosome 7 open reading frame 55 |
| chr6_-_111927062 | 0.60 |
ENST00000359831.4
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr17_+_43239191 | 0.58 |
ENST00000589230.1
|
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr11_-_111649015 | 0.57 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr10_+_94594351 | 0.57 |
ENST00000371552.4
|
EXOC6
|
exocyst complex component 6 |
| chr12_+_107712173 | 0.56 |
ENST00000280758.5
ENST00000420571.2 |
BTBD11
|
BTB (POZ) domain containing 11 |
| chr6_+_12290586 | 0.56 |
ENST00000379375.5
|
EDN1
|
endothelin 1 |
| chr18_-_24237339 | 0.55 |
ENST00000580191.1
|
KCTD1
|
potassium channel tetramerization domain containing 1 |
| chr17_+_17685422 | 0.53 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1 |
| chr22_+_42834029 | 0.50 |
ENST00000428765.1
|
CTA-126B4.7
|
CTA-126B4.7 |
| chr17_+_43239231 | 0.49 |
ENST00000591576.1
ENST00000591070.1 ENST00000592695.1 |
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr4_+_129732419 | 0.49 |
ENST00000510308.1
|
PHF17
|
jade family PHD finger 1 |
| chr7_-_81635106 | 0.48 |
ENST00000443883.1
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chr3_-_27764190 | 0.46 |
ENST00000537516.1
|
EOMES
|
eomesodermin |
| chr6_-_97731019 | 0.44 |
ENST00000275053.4
|
MMS22L
|
MMS22-like, DNA repair protein |
| chr8_+_38831683 | 0.43 |
ENST00000302495.4
|
HTRA4
|
HtrA serine peptidase 4 |
| chr4_+_129732467 | 0.42 |
ENST00000413543.2
|
PHF17
|
jade family PHD finger 1 |
| chr8_+_107738343 | 0.41 |
ENST00000521592.1
|
OXR1
|
oxidation resistance 1 |
| chr11_-_31531121 | 0.41 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr12_-_102591604 | 0.40 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr17_+_59489112 | 0.40 |
ENST00000335108.2
|
C17orf82
|
chromosome 17 open reading frame 82 |
| chr4_-_84035868 | 0.38 |
ENST00000426923.2
ENST00000509973.1 |
PLAC8
|
placenta-specific 8 |
| chrX_+_55478538 | 0.37 |
ENST00000342972.1
|
MAGEH1
|
melanoma antigen family H, 1 |
| chr9_+_116225999 | 0.37 |
ENST00000317613.6
|
RGS3
|
regulator of G-protein signaling 3 |
| chr14_+_65170820 | 0.36 |
ENST00000555982.1
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr5_+_60933634 | 0.36 |
ENST00000505642.1
|
C5orf64
|
chromosome 5 open reading frame 64 |
| chr8_+_26150628 | 0.35 |
ENST00000523925.1
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr12_-_2113583 | 0.34 |
ENST00000397173.4
ENST00000280665.6 |
DCP1B
|
decapping mRNA 1B |
| chr1_+_226411319 | 0.34 |
ENST00000542034.1
ENST00000366810.5 |
MIXL1
|
Mix paired-like homeobox |
| chr7_-_107880508 | 0.33 |
ENST00000425651.2
|
NRCAM
|
neuronal cell adhesion molecule |
| chr2_-_224467093 | 0.33 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chrX_-_130423240 | 0.32 |
ENST00000370910.1
ENST00000370901.4 |
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr14_-_82000140 | 0.31 |
ENST00000555824.1
ENST00000557372.1 ENST00000336735.4 |
SEL1L
|
sel-1 suppressor of lin-12-like (C. elegans) |
| chr6_+_130339710 | 0.30 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr12_-_53994805 | 0.30 |
ENST00000328463.7
|
ATF7
|
activating transcription factor 7 |
| chr12_-_57328187 | 0.29 |
ENST00000293502.1
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chr14_-_27066636 | 0.29 |
ENST00000267422.7
ENST00000344429.5 ENST00000574031.1 ENST00000465357.2 ENST00000547619.1 |
NOVA1
|
neuro-oncological ventral antigen 1 |
| chr4_+_37455536 | 0.29 |
ENST00000381980.4
ENST00000508175.1 |
C4orf19
|
chromosome 4 open reading frame 19 |
| chr2_-_203776864 | 0.28 |
ENST00000261015.4
|
WDR12
|
WD repeat domain 12 |
| chr18_-_74207146 | 0.28 |
ENST00000443185.2
|
ZNF516
|
zinc finger protein 516 |
| chr17_+_44701402 | 0.27 |
ENST00000575068.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr16_-_67517716 | 0.27 |
ENST00000290953.2
|
AGRP
|
agouti related protein homolog (mouse) |
| chr17_+_68101117 | 0.26 |
ENST00000587698.1
ENST00000587892.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr6_-_161695074 | 0.26 |
ENST00000457520.2
ENST00000366906.5 ENST00000320285.4 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr1_+_43291220 | 0.26 |
ENST00000372514.3
|
ERMAP
|
erythroblast membrane-associated protein (Scianna blood group) |
| chrX_+_123097014 | 0.25 |
ENST00000394478.1
|
STAG2
|
stromal antigen 2 |
| chr2_-_164592497 | 0.24 |
ENST00000333129.3
ENST00000409634.1 |
FIGN
|
fidgetin |
| chr2_+_203776937 | 0.24 |
ENST00000402905.3
ENST00000414490.1 ENST00000431787.1 ENST00000444724.1 ENST00000414857.1 ENST00000430899.1 ENST00000445120.1 ENST00000441569.1 ENST00000432024.1 ENST00000443740.1 ENST00000414439.1 ENST00000428585.1 ENST00000545253.1 ENST00000545262.1 ENST00000447539.1 ENST00000456821.2 ENST00000434998.1 ENST00000320443.8 |
CARF
|
calcium responsive transcription factor |
| chr10_+_74451883 | 0.23 |
ENST00000373053.3
ENST00000357157.6 |
MCU
|
mitochondrial calcium uniporter |
| chr7_-_104909435 | 0.22 |
ENST00000357311.3
|
SRPK2
|
SRSF protein kinase 2 |
| chr12_-_122018346 | 0.22 |
ENST00000377069.4
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr13_+_76378305 | 0.22 |
ENST00000526371.1
ENST00000526528.1 |
LMO7
|
LIM domain 7 |
| chr18_+_20714525 | 0.22 |
ENST00000400473.2
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1 |
| chr17_-_46716647 | 0.21 |
ENST00000608940.1
|
RP11-357H14.17
|
RP11-357H14.17 |
| chr13_-_26795840 | 0.20 |
ENST00000381570.3
ENST00000399762.2 ENST00000346166.3 |
RNF6
|
ring finger protein (C3H2C3 type) 6 |
| chr14_+_32798547 | 0.20 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr17_+_47448102 | 0.20 |
ENST00000576461.1
|
RP11-81K2.1
|
Uncharacterized protein |
| chr5_-_142780280 | 0.19 |
ENST00000424646.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr12_-_51611477 | 0.19 |
ENST00000389243.4
|
POU6F1
|
POU class 6 homeobox 1 |
| chr6_+_101846664 | 0.19 |
ENST00000421544.1
ENST00000413795.1 ENST00000369138.1 ENST00000358361.3 |
GRIK2
|
glutamate receptor, ionotropic, kainate 2 |
| chr3_+_130569429 | 0.18 |
ENST00000505330.1
ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr16_-_49890016 | 0.18 |
ENST00000563137.2
|
ZNF423
|
zinc finger protein 423 |
| chr9_+_136501478 | 0.17 |
ENST00000393056.2
ENST00000263611.2 |
DBH
|
dopamine beta-hydroxylase (dopamine beta-monooxygenase) |
| chr16_+_70207686 | 0.16 |
ENST00000541793.2
ENST00000314151.8 ENST00000565806.1 ENST00000569347.2 ENST00000536907.2 |
CLEC18C
|
C-type lectin domain family 18, member C |
| chr1_+_150337144 | 0.15 |
ENST00000539519.1
ENST00000369067.3 ENST00000369068.4 |
RPRD2
|
regulation of nuclear pre-mRNA domain containing 2 |
| chr20_-_50722183 | 0.15 |
ENST00000371523.4
|
ZFP64
|
ZFP64 zinc finger protein |
| chr2_-_165698662 | 0.15 |
ENST00000194871.6
ENST00000445474.2 |
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chrX_-_46187069 | 0.15 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr6_-_32157947 | 0.14 |
ENST00000375050.4
|
PBX2
|
pre-B-cell leukemia homeobox 2 |
| chr10_-_13350447 | 0.13 |
ENST00000429930.1
|
AL138764.1
|
Uncharacterized protein |
| chr1_+_153746683 | 0.13 |
ENST00000271857.2
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chrX_+_133733457 | 0.12 |
ENST00000440614.1
|
RP11-308B5.2
|
RP11-308B5.2 |
| chr7_+_18535346 | 0.12 |
ENST00000405010.3
ENST00000406451.4 ENST00000428307.2 |
HDAC9
|
histone deacetylase 9 |
| chr12_-_94673956 | 0.12 |
ENST00000551941.1
|
RP11-1105G2.3
|
Uncharacterized protein |
| chr12_-_122017542 | 0.12 |
ENST00000446152.2
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr11_+_123430948 | 0.12 |
ENST00000529432.1
ENST00000534764.1 |
GRAMD1B
|
GRAM domain containing 1B |
| chr7_+_148982396 | 0.11 |
ENST00000418158.2
|
ZNF783
|
zinc finger family member 783 |
| chr7_+_18535321 | 0.11 |
ENST00000413380.1
ENST00000430454.1 |
HDAC9
|
histone deacetylase 9 |
| chr6_-_76203345 | 0.11 |
ENST00000393004.2
|
FILIP1
|
filamin A interacting protein 1 |
| chr12_-_86650077 | 0.11 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr18_-_21977748 | 0.11 |
ENST00000399441.4
ENST00000319481.3 |
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr2_-_165698521 | 0.11 |
ENST00000409184.3
ENST00000392717.2 ENST00000456693.1 |
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr16_+_69985083 | 0.11 |
ENST00000288040.6
ENST00000449317.2 |
CLEC18A
|
C-type lectin domain family 18, member A |
| chr4_+_86749045 | 0.11 |
ENST00000514229.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr5_+_61874562 | 0.11 |
ENST00000334994.5
ENST00000409534.1 |
LRRC70
IPO11
|
leucine rich repeat containing 70 importin 11 |
| chr6_+_114178512 | 0.11 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr4_-_84035905 | 0.10 |
ENST00000311507.4
|
PLAC8
|
placenta-specific 8 |
| chrX_+_105937068 | 0.09 |
ENST00000324342.3
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr1_+_150337100 | 0.09 |
ENST00000401000.4
|
RPRD2
|
regulation of nuclear pre-mRNA domain containing 2 |
| chr11_-_72070206 | 0.09 |
ENST00000544382.1
|
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr18_-_53253112 | 0.09 |
ENST00000568673.1
ENST00000562847.1 ENST00000568147.1 |
TCF4
|
transcription factor 4 |
| chr4_+_86525299 | 0.09 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr5_+_65222500 | 0.09 |
ENST00000511297.1
ENST00000506030.1 |
ERBB2IP
|
erbb2 interacting protein |
| chr13_-_28545276 | 0.09 |
ENST00000381020.7
|
CDX2
|
caudal type homeobox 2 |
| chr16_-_74455290 | 0.09 |
ENST00000339953.5
|
CLEC18B
|
C-type lectin domain family 18, member B |
| chr8_+_97773202 | 0.08 |
ENST00000519484.1
|
CPQ
|
carboxypeptidase Q |
| chr19_-_49622348 | 0.08 |
ENST00000408991.2
|
C19orf73
|
chromosome 19 open reading frame 73 |
| chr6_-_138820624 | 0.08 |
ENST00000343505.5
|
NHSL1
|
NHS-like 1 |
| chr8_-_109799793 | 0.08 |
ENST00000297459.3
|
TMEM74
|
transmembrane protein 74 |
| chr12_-_12674032 | 0.08 |
ENST00000298573.4
|
DUSP16
|
dual specificity phosphatase 16 |
| chr6_-_139613269 | 0.07 |
ENST00000358430.3
|
TXLNB
|
taxilin beta |
| chr9_+_109685630 | 0.07 |
ENST00000451160.2
|
RP11-508N12.4
|
Uncharacterized protein |
| chr12_-_120241187 | 0.07 |
ENST00000392520.2
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr1_-_144994909 | 0.06 |
ENST00000369347.4
ENST00000369354.3 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr6_-_76203454 | 0.06 |
ENST00000237172.7
|
FILIP1
|
filamin A interacting protein 1 |
| chr11_-_117170403 | 0.06 |
ENST00000504995.1
|
BACE1
|
beta-site APP-cleaving enzyme 1 |
| chr4_-_122148620 | 0.06 |
ENST00000509841.1
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr1_+_62439037 | 0.06 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr2_+_135596180 | 0.06 |
ENST00000283054.4
ENST00000392928.1 |
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr11_+_46722368 | 0.05 |
ENST00000311764.2
|
ZNF408
|
zinc finger protein 408 |
| chr16_-_51185172 | 0.05 |
ENST00000251020.4
|
SALL1
|
spalt-like transcription factor 1 |
| chr17_+_39845134 | 0.05 |
ENST00000591776.1
ENST00000469257.1 |
EIF1
|
eukaryotic translation initiation factor 1 |
| chr1_+_244515930 | 0.05 |
ENST00000366537.1
ENST00000308105.4 |
C1orf100
|
chromosome 1 open reading frame 100 |
| chr18_+_55888767 | 0.04 |
ENST00000431212.2
ENST00000586268.1 ENST00000587190.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr11_+_35198118 | 0.04 |
ENST00000525211.1
ENST00000526000.1 ENST00000279452.6 ENST00000527889.1 |
CD44
|
CD44 molecule (Indian blood group) |
| chrX_-_130423200 | 0.04 |
ENST00000361420.3
|
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr4_+_71587669 | 0.04 |
ENST00000381006.3
ENST00000226328.4 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr21_+_39644395 | 0.03 |
ENST00000398934.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr10_-_62332357 | 0.03 |
ENST00000503366.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr1_+_40713573 | 0.03 |
ENST00000372766.3
|
TMCO2
|
transmembrane and coiled-coil domains 2 |
| chr5_+_43121698 | 0.03 |
ENST00000505606.2
ENST00000509634.1 ENST00000509341.1 |
ZNF131
|
zinc finger protein 131 |
| chr12_-_101604185 | 0.03 |
ENST00000536262.2
|
SLC5A8
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 8 |
| chr11_+_64018955 | 0.02 |
ENST00000279230.6
ENST00000540288.1 ENST00000325234.5 |
PLCB3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chrX_-_21676442 | 0.02 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr16_+_69984810 | 0.02 |
ENST00000393701.2
ENST00000568461.1 |
CLEC18A
|
C-type lectin domain family 18, member A |
| chr8_-_103425047 | 0.02 |
ENST00000520539.1
|
UBR5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr4_+_169418255 | 0.02 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr8_-_103424986 | 0.02 |
ENST00000521922.1
|
UBR5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr1_-_95391315 | 0.00 |
ENST00000545882.1
ENST00000415017.1 |
CNN3
|
calponin 3, acidic |
Network of associatons between targets according to the STRING database.
First level regulatory network of VENTX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.2 | 0.6 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.1 | 1.7 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.7 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.1 | 0.7 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.5 | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.1 | 1.0 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 1.8 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 0.3 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.1 | 0.5 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.1 | 0.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.3 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 0.2 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 1.2 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.4 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 1.0 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 1.2 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.2 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.4 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.3 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.3 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.9 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.3 | GO:0008343 | adult feeding behavior(GO:0008343) positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.2 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.0 | 0.7 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.9 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.3 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.9 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:0045209 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.3 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.3 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.9 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.4 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 0.6 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 1.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 0.4 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 1.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.3 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.0 | 0.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.2 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 1.4 | GO:0035578 | azurophil granule lumen(GO:0035578) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.2 | 0.5 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.1 | 0.6 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 1.4 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 1.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 1.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.4 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.2 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 1.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.3 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 1.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.7 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 1.0 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 1.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.9 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |