Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
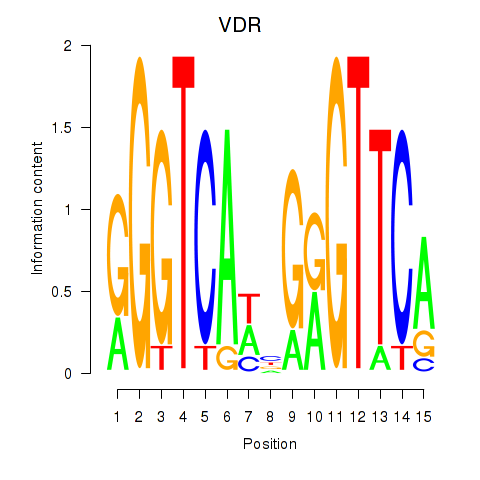
Results for VDR
Z-value: 1.31
Transcription factors associated with VDR
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
VDR
|
ENSG00000111424.6 | vitamin D receptor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| VDR | hg19_v2_chr12_-_48298785_48298828 | 0.68 | 3.2e-01 | Click! |
Activity profile of VDR motif
Sorted Z-values of VDR motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_17579110 | 0.93 |
ENST00000606142.1
|
LAP3
|
leucine aminopeptidase 3 |
| chr11_-_60719213 | 0.87 |
ENST00000227880.3
|
SLC15A3
|
solute carrier family 15 (oligopeptide transporter), member 3 |
| chr11_+_65154070 | 0.81 |
ENST00000317568.5
ENST00000531296.1 ENST00000533782.1 ENST00000355991.5 ENST00000416776.2 ENST00000526201.1 |
FRMD8
|
FERM domain containing 8 |
| chr10_+_35484793 | 0.81 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr6_+_31916733 | 0.80 |
ENST00000483004.1
|
CFB
|
complement factor B |
| chr1_-_200992827 | 0.77 |
ENST00000332129.2
ENST00000422435.2 |
KIF21B
|
kinesin family member 21B |
| chr1_+_200993071 | 0.75 |
ENST00000446333.1
ENST00000458003.1 |
RP11-168O16.1
|
RP11-168O16.1 |
| chr12_+_52626898 | 0.74 |
ENST00000331817.5
|
KRT7
|
keratin 7 |
| chr5_-_147211226 | 0.65 |
ENST00000296695.5
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1 |
| chr7_-_92777606 | 0.62 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr6_+_46661575 | 0.62 |
ENST00000450697.1
|
TDRD6
|
tudor domain containing 6 |
| chr5_-_177209832 | 0.61 |
ENST00000393518.3
ENST00000505531.1 ENST00000503567.1 |
FAM153A
|
family with sequence similarity 153, member A |
| chr12_+_52695617 | 0.60 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chr9_+_139839711 | 0.60 |
ENST00000224181.3
|
C8G
|
complement component 8, gamma polypeptide |
| chr1_-_183559693 | 0.60 |
ENST00000367535.3
ENST00000413720.1 ENST00000418089.1 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr2_+_169926047 | 0.60 |
ENST00000428522.1
ENST00000450153.1 ENST00000421653.1 |
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr12_+_100967533 | 0.58 |
ENST00000550295.1
|
GAS2L3
|
growth arrest-specific 2 like 3 |
| chr2_+_231280954 | 0.53 |
ENST00000409824.1
ENST00000409341.1 ENST00000409112.1 ENST00000340126.4 ENST00000341950.4 |
SP100
|
SP100 nuclear antigen |
| chr2_+_233527443 | 0.51 |
ENST00000410095.1
|
EFHD1
|
EF-hand domain family, member D1 |
| chr11_-_104817919 | 0.51 |
ENST00000533252.1
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr20_+_3801162 | 0.50 |
ENST00000379573.2
ENST00000379567.2 ENST00000455742.1 ENST00000246041.2 |
AP5S1
|
adaptor-related protein complex 5, sigma 1 subunit |
| chr12_+_27091387 | 0.50 |
ENST00000544111.1
|
FGFR1OP2
|
FGFR1 oncogene partner 2 |
| chr14_+_52780998 | 0.49 |
ENST00000557436.1
|
PTGER2
|
prostaglandin E receptor 2 (subtype EP2), 53kDa |
| chr15_-_75230478 | 0.46 |
ENST00000322347.6
|
COX5A
|
cytochrome c oxidase subunit Va |
| chr16_+_19078960 | 0.46 |
ENST00000568985.1
ENST00000566110.1 |
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr8_+_119294456 | 0.45 |
ENST00000366457.2
|
AC023590.1
|
Uncharacterized protein |
| chr2_+_210518057 | 0.45 |
ENST00000452717.1
|
MAP2
|
microtubule-associated protein 2 |
| chr15_-_75230368 | 0.44 |
ENST00000564811.1
ENST00000562233.1 ENST00000567270.1 ENST00000568783.1 |
COX5A
|
cytochrome c oxidase subunit Va |
| chr3_-_122283079 | 0.44 |
ENST00000471785.1
ENST00000466126.1 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr7_+_129015484 | 0.44 |
ENST00000490911.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr3_-_122283100 | 0.43 |
ENST00000492382.1
ENST00000462315.1 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr1_+_2487078 | 0.43 |
ENST00000426449.1
ENST00000434817.1 ENST00000435221.2 |
TNFRSF14
|
tumor necrosis factor receptor superfamily, member 14 |
| chr5_+_175487692 | 0.43 |
ENST00000510151.1
|
FAM153B
|
family with sequence similarity 153, member B |
| chr12_-_58135903 | 0.42 |
ENST00000257897.3
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr1_+_92414952 | 0.42 |
ENST00000449584.1
ENST00000427104.1 ENST00000355011.3 ENST00000448194.1 ENST00000426141.1 ENST00000450792.1 ENST00000548992.1 ENST00000552654.1 ENST00000457265.1 |
BRDT
|
bromodomain, testis-specific |
| chr2_+_210517895 | 0.41 |
ENST00000447185.1
|
MAP2
|
microtubule-associated protein 2 |
| chr21_+_45905448 | 0.40 |
ENST00000449713.1
|
AP001065.15
|
AP001065.15 |
| chr8_-_135522425 | 0.40 |
ENST00000521673.1
|
ZFAT
|
zinc finger and AT hook domain containing |
| chr14_-_62217779 | 0.40 |
ENST00000554254.1
|
HIF1A-AS2
|
HIF1A antisense RNA 2 |
| chr11_+_57365150 | 0.40 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr10_+_104154229 | 0.39 |
ENST00000428099.1
ENST00000369966.3 |
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr2_+_231280908 | 0.39 |
ENST00000427101.2
ENST00000432979.1 |
SP100
|
SP100 nuclear antigen |
| chr11_+_82783097 | 0.39 |
ENST00000501011.2
ENST00000527627.1 ENST00000526795.1 ENST00000533528.1 ENST00000533708.1 ENST00000534499.1 |
RAB30-AS1
|
RAB30 antisense RNA 1 (head to head) |
| chr1_-_224624730 | 0.39 |
ENST00000445239.1
|
WDR26
|
WD repeat domain 26 |
| chr8_-_23563922 | 0.39 |
ENST00000418222.1
ENST00000325017.3 |
NKX2-6
|
NK2 homeobox 6 |
| chr16_+_19079215 | 0.38 |
ENST00000544894.2
ENST00000561858.1 |
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr6_+_43603552 | 0.37 |
ENST00000372171.4
|
MAD2L1BP
|
MAD2L1 binding protein |
| chr12_-_53575120 | 0.37 |
ENST00000542115.1
|
CSAD
|
cysteine sulfinic acid decarboxylase |
| chr1_+_47137445 | 0.37 |
ENST00000569393.1
ENST00000334122.4 ENST00000415500.1 |
TEX38
|
testis expressed 38 |
| chr12_+_110011571 | 0.37 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr9_-_139838986 | 0.35 |
ENST00000443788.1
|
FBXW5
|
F-box and WD repeat domain containing 5 |
| chr14_+_101295948 | 0.35 |
ENST00000452514.2
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr18_+_20714525 | 0.33 |
ENST00000400473.2
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1 |
| chr3_+_48264816 | 0.33 |
ENST00000296435.2
ENST00000576243.1 |
CAMP
|
cathelicidin antimicrobial peptide |
| chr19_+_54694119 | 0.33 |
ENST00000456872.1
ENST00000302937.4 ENST00000429671.2 |
TSEN34
|
TSEN34 tRNA splicing endonuclease subunit |
| chr16_+_14802801 | 0.33 |
ENST00000526520.1
ENST00000531598.2 |
NPIPA3
|
nuclear pore complex interacting protein family, member A3 |
| chr12_-_109221160 | 0.33 |
ENST00000326470.5
|
SSH1
|
slingshot protein phosphatase 1 |
| chr7_+_99699280 | 0.32 |
ENST00000421755.1
|
AP4M1
|
adaptor-related protein complex 4, mu 1 subunit |
| chr7_+_120590803 | 0.32 |
ENST00000315870.5
ENST00000339121.5 ENST00000445699.1 |
ING3
|
inhibitor of growth family, member 3 |
| chr1_+_175036966 | 0.31 |
ENST00000239462.4
|
TNN
|
tenascin N |
| chr11_-_61582579 | 0.31 |
ENST00000539419.1
ENST00000545245.1 ENST00000545405.1 ENST00000542506.1 |
FADS1
|
fatty acid desaturase 1 |
| chr3_+_184033551 | 0.30 |
ENST00000456033.1
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr19_+_589893 | 0.30 |
ENST00000251287.2
|
HCN2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2 |
| chr17_-_42992856 | 0.30 |
ENST00000588316.1
ENST00000435360.2 ENST00000586793.1 ENST00000588735.1 ENST00000588037.1 ENST00000592320.1 ENST00000253408.5 |
GFAP
|
glial fibrillary acidic protein |
| chr8_-_93115445 | 0.30 |
ENST00000523629.1
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr17_-_76778339 | 0.30 |
ENST00000591455.1
ENST00000446868.3 ENST00000361101.4 ENST00000589296.1 |
CYTH1
|
cytohesin 1 |
| chr2_+_87769459 | 0.30 |
ENST00000414030.1
ENST00000437561.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr10_+_47894572 | 0.30 |
ENST00000355876.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr20_+_36974759 | 0.30 |
ENST00000217407.2
|
LBP
|
lipopolysaccharide binding protein |
| chr7_-_45960850 | 0.29 |
ENST00000381083.4
ENST00000381086.5 ENST00000275521.6 |
IGFBP3
|
insulin-like growth factor binding protein 3 |
| chr11_+_124609823 | 0.29 |
ENST00000412681.2
|
NRGN
|
neurogranin (protein kinase C substrate, RC3) |
| chr19_-_47987419 | 0.29 |
ENST00000536339.1
ENST00000595554.1 ENST00000600271.1 ENST00000338134.3 |
KPTN
|
kaptin (actin binding protein) |
| chr17_-_36347835 | 0.29 |
ENST00000519532.1
|
TBC1D3
|
TBC1 domain family, member 3 |
| chr17_-_34807272 | 0.29 |
ENST00000535592.1
ENST00000394453.1 |
TBC1D3G
|
TBC1 domain family, member 3G |
| chr17_-_34756219 | 0.29 |
ENST00000451448.2
ENST00000394359.3 |
TBC1D3C
TBC1D3H
|
TBC1 domain family, member 3C TBC1 domain family, member 3H |
| chr7_+_143078379 | 0.29 |
ENST00000449630.1
ENST00000457235.1 |
ZYX
|
zyxin |
| chr11_+_6624970 | 0.29 |
ENST00000420936.2
ENST00000528995.1 |
ILK
|
integrin-linked kinase |
| chr19_+_56652686 | 0.28 |
ENST00000592949.1
|
ZNF444
|
zinc finger protein 444 |
| chr6_+_13272904 | 0.28 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr19_+_38826415 | 0.27 |
ENST00000410018.1
ENST00000409235.3 |
CATSPERG
|
catsper channel auxiliary subunit gamma |
| chr16_+_16043406 | 0.27 |
ENST00000399410.3
ENST00000399408.2 ENST00000346370.5 ENST00000351154.5 ENST00000345148.5 ENST00000349029.5 |
ABCC1
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
| chr8_+_23104130 | 0.27 |
ENST00000313219.7
ENST00000519984.1 |
CHMP7
|
charged multivesicular body protein 7 |
| chr17_-_40288449 | 0.27 |
ENST00000552162.1
ENST00000550504.1 |
RAB5C
|
RAB5C, member RAS oncogene family |
| chr5_-_57854070 | 0.26 |
ENST00000504333.1
|
CTD-2117L12.1
|
Uncharacterized protein |
| chr3_-_71114066 | 0.26 |
ENST00000485326.2
|
FOXP1
|
forkhead box P1 |
| chr11_+_6624955 | 0.26 |
ENST00000299421.4
ENST00000537806.1 |
ILK
|
integrin-linked kinase |
| chr17_+_61473104 | 0.26 |
ENST00000583016.1
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr2_+_176957619 | 0.26 |
ENST00000392539.3
|
HOXD13
|
homeobox D13 |
| chr16_+_31044812 | 0.25 |
ENST00000313843.3
|
STX4
|
syntaxin 4 |
| chr1_+_2487402 | 0.25 |
ENST00000451778.1
|
TNFRSF14
|
tumor necrosis factor receptor superfamily, member 14 |
| chr13_-_33859819 | 0.25 |
ENST00000336934.5
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr17_-_34808047 | 0.25 |
ENST00000592614.1
ENST00000591542.1 ENST00000330458.7 ENST00000341264.6 ENST00000592987.1 ENST00000400684.4 |
TBC1D3G
TBC1D3H
|
TBC1 domain family, member 3G TBC1 domain family, member 3H |
| chr7_-_151433393 | 0.25 |
ENST00000492843.1
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr22_+_35776354 | 0.25 |
ENST00000412893.1
|
HMOX1
|
heme oxygenase (decycling) 1 |
| chr19_-_54693401 | 0.25 |
ENST00000338624.6
|
MBOAT7
|
membrane bound O-acyltransferase domain containing 7 |
| chr5_+_177433973 | 0.25 |
ENST00000507848.1
|
FAM153C
|
family with sequence similarity 153, member C |
| chr7_-_151433342 | 0.25 |
ENST00000433631.2
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr7_+_102295340 | 0.24 |
ENST00000455020.2
|
SPDYE2B
|
speedy/RINGO cell cycle regulator family member E2B |
| chr14_-_35873856 | 0.24 |
ENST00000553342.1
ENST00000216797.5 ENST00000557140.1 |
NFKBIA
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
| chr14_-_23398565 | 0.24 |
ENST00000397440.4
ENST00000538452.1 ENST00000421938.2 ENST00000554867.1 ENST00000556616.1 ENST00000216350.8 ENST00000553550.1 ENST00000397441.2 ENST00000553897.1 |
PRMT5
|
protein arginine methyltransferase 5 |
| chr19_-_5903714 | 0.24 |
ENST00000586349.1
ENST00000585661.1 ENST00000308961.4 ENST00000592634.1 ENST00000418389.2 ENST00000252675.5 |
AC024592.12
NDUFA11
FUT5
|
Uncharacterized protein NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa fucosyltransferase 5 (alpha (1,3) fucosyltransferase) |
| chr6_-_30524951 | 0.24 |
ENST00000376621.3
|
GNL1
|
guanine nucleotide binding protein-like 1 |
| chr16_+_57392684 | 0.24 |
ENST00000219235.4
|
CCL22
|
chemokine (C-C motif) ligand 22 |
| chr11_-_64527425 | 0.24 |
ENST00000377432.3
|
PYGM
|
phosphorylase, glycogen, muscle |
| chr5_-_131630931 | 0.24 |
ENST00000431054.1
|
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chr6_+_30853002 | 0.24 |
ENST00000421124.2
ENST00000512725.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr3_+_184032919 | 0.24 |
ENST00000427845.1
ENST00000342981.4 ENST00000319274.6 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr17_+_36284791 | 0.23 |
ENST00000505415.1
|
TBC1D3F
|
TBC1 domain family, member 3F |
| chr2_-_230786619 | 0.23 |
ENST00000389045.3
ENST00000409677.1 |
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr16_-_87799505 | 0.23 |
ENST00000353170.5
ENST00000561825.1 ENST00000270583.5 ENST00000562261.1 ENST00000347925.5 |
KLHDC4
|
kelch domain containing 4 |
| chr16_+_19079311 | 0.23 |
ENST00000569127.1
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr12_+_56552128 | 0.23 |
ENST00000548580.1
ENST00000293422.5 ENST00000348108.4 ENST00000549017.1 ENST00000549566.1 ENST00000536128.1 ENST00000547649.1 ENST00000547408.1 ENST00000551589.1 ENST00000549392.1 ENST00000548400.1 ENST00000548293.1 |
MYL6
|
myosin, light chain 6, alkali, smooth muscle and non-muscle |
| chr3_+_122283064 | 0.23 |
ENST00000296161.4
|
DTX3L
|
deltex 3-like (Drosophila) |
| chr22_-_29663954 | 0.22 |
ENST00000216085.7
|
RHBDD3
|
rhomboid domain containing 3 |
| chr18_+_9102669 | 0.22 |
ENST00000497577.2
|
NDUFV2
|
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
| chr12_+_57610562 | 0.22 |
ENST00000349394.5
|
NXPH4
|
neurexophilin 4 |
| chr17_-_36348610 | 0.22 |
ENST00000339023.4
ENST00000354664.4 |
TBC1D3
|
TBC1 domain family, member 3 |
| chr3_+_54157480 | 0.22 |
ENST00000490478.1
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chr8_+_95653373 | 0.22 |
ENST00000358397.5
|
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr1_-_19426149 | 0.22 |
ENST00000429347.2
|
UBR4
|
ubiquitin protein ligase E3 component n-recognin 4 |
| chr12_+_3068957 | 0.22 |
ENST00000543035.1
|
TEAD4
|
TEA domain family member 4 |
| chr17_-_79791118 | 0.22 |
ENST00000576431.1
ENST00000575061.1 ENST00000455127.2 ENST00000572645.1 ENST00000538396.1 ENST00000573478.1 |
FAM195B
|
family with sequence similarity 195, member B |
| chr6_-_32557610 | 0.22 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr11_-_4629367 | 0.21 |
ENST00000533021.1
|
TRIM68
|
tripartite motif containing 68 |
| chrX_+_122993827 | 0.21 |
ENST00000371199.3
|
XIAP
|
X-linked inhibitor of apoptosis |
| chr1_+_47137544 | 0.21 |
ENST00000564373.1
|
TEX38
|
testis expressed 38 |
| chr16_+_19078911 | 0.21 |
ENST00000321998.5
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr16_-_31105870 | 0.21 |
ENST00000394971.3
|
VKORC1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr14_+_95047744 | 0.20 |
ENST00000553511.1
ENST00000554633.1 ENST00000555681.1 ENST00000554276.1 |
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr19_+_18077881 | 0.20 |
ENST00000609922.1
|
KCNN1
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 |
| chr19_+_39936267 | 0.20 |
ENST00000359191.6
|
SUPT5H
|
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr19_-_13261160 | 0.20 |
ENST00000343587.5
ENST00000591197.1 |
STX10
|
syntaxin 10 |
| chr11_+_6625046 | 0.20 |
ENST00000396751.2
|
ILK
|
integrin-linked kinase |
| chr5_-_177210399 | 0.20 |
ENST00000510276.1
|
FAM153A
|
family with sequence similarity 153, member A |
| chr19_-_49122384 | 0.20 |
ENST00000550973.1
ENST00000550645.1 ENST00000549273.1 ENST00000552588.1 |
RPL18
|
ribosomal protein L18 |
| chr16_-_31106048 | 0.20 |
ENST00000300851.6
|
VKORC1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr16_-_31106211 | 0.20 |
ENST00000532364.1
ENST00000529564.1 ENST00000319788.7 ENST00000354895.4 ENST00000394975.2 |
RP11-196G11.1
VKORC1
|
Uncharacterized protein vitamin K epoxide reductase complex, subunit 1 |
| chr19_-_13261090 | 0.19 |
ENST00000588848.1
|
STX10
|
syntaxin 10 |
| chrX_+_100645812 | 0.19 |
ENST00000427805.2
ENST00000553110.3 ENST00000392994.3 ENST00000409338.1 ENST00000409170.3 |
RPL36A
RPL36A-HNRNPH2
|
ribosomal protein L36a RPL36A-HNRNPH2 readthrough |
| chr3_-_112218378 | 0.19 |
ENST00000334529.5
|
BTLA
|
B and T lymphocyte associated |
| chr15_+_83776324 | 0.19 |
ENST00000379390.6
ENST00000379386.4 ENST00000565774.1 ENST00000565982.1 |
TM6SF1
|
transmembrane 6 superfamily member 1 |
| chr19_+_57831829 | 0.19 |
ENST00000321545.4
|
ZNF543
|
zinc finger protein 543 |
| chr10_+_99627889 | 0.19 |
ENST00000596005.1
|
GOLGA7B
|
Golgin subfamily A member 7B; cDNA FLJ43465 fis, clone OCBBF2036476 |
| chr1_-_6550625 | 0.19 |
ENST00000377725.1
ENST00000340850.5 |
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr19_+_41281282 | 0.19 |
ENST00000263369.3
|
MIA
|
melanoma inhibitory activity |
| chr19_-_11494975 | 0.19 |
ENST00000222139.6
ENST00000592375.2 |
EPOR
|
erythropoietin receptor |
| chr11_-_73882029 | 0.19 |
ENST00000539061.1
|
C2CD3
|
C2 calcium-dependent domain containing 3 |
| chr11_-_104840093 | 0.19 |
ENST00000417440.2
ENST00000444739.2 |
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr19_-_12595586 | 0.18 |
ENST00000397732.3
|
ZNF709
|
zinc finger protein 709 |
| chr1_-_24741525 | 0.18 |
ENST00000374409.1
|
STPG1
|
sperm-tail PG-rich repeat containing 1 |
| chr3_+_184033135 | 0.18 |
ENST00000424196.1
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr15_+_75940218 | 0.18 |
ENST00000308527.5
|
SNX33
|
sorting nexin 33 |
| chr4_-_165898768 | 0.18 |
ENST00000329314.5
|
TRIM61
|
tripartite motif containing 61 |
| chr3_+_119298280 | 0.18 |
ENST00000481816.1
|
ADPRH
|
ADP-ribosylarginine hydrolase |
| chr18_+_42277095 | 0.18 |
ENST00000591940.1
|
SETBP1
|
SET binding protein 1 |
| chr16_-_67281413 | 0.18 |
ENST00000258201.4
|
FHOD1
|
formin homology 2 domain containing 1 |
| chr1_+_154474689 | 0.18 |
ENST00000368482.4
|
TDRD10
|
tudor domain containing 10 |
| chr6_+_56820152 | 0.17 |
ENST00000370745.1
|
BEND6
|
BEN domain containing 6 |
| chr16_-_75282088 | 0.17 |
ENST00000542031.2
|
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr7_-_107770794 | 0.17 |
ENST00000205386.4
ENST00000418464.1 ENST00000388781.3 ENST00000388780.3 ENST00000414450.2 |
LAMB4
|
laminin, beta 4 |
| chr2_-_203736334 | 0.17 |
ENST00000392237.2
ENST00000416760.1 ENST00000412210.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr1_+_198608146 | 0.17 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr19_+_39936186 | 0.17 |
ENST00000432763.2
ENST00000402194.2 ENST00000601515.1 |
SUPT5H
|
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr11_-_65667884 | 0.17 |
ENST00000448083.2
ENST00000531493.1 ENST00000532401.1 |
FOSL1
|
FOS-like antigen 1 |
| chr12_+_56522001 | 0.17 |
ENST00000267113.4
ENST00000541590.1 |
ESYT1
|
extended synaptotagmin-like protein 1 |
| chr13_+_47127322 | 0.17 |
ENST00000389798.3
|
LRCH1
|
leucine-rich repeats and calponin homology (CH) domain containing 1 |
| chr16_+_532503 | 0.17 |
ENST00000412256.1
|
RAB11FIP3
|
RAB11 family interacting protein 3 (class II) |
| chr12_+_48178706 | 0.17 |
ENST00000599515.1
|
AC004466.1
|
Uncharacterized protein |
| chr3_+_46618727 | 0.17 |
ENST00000296145.5
|
TDGF1
|
teratocarcinoma-derived growth factor 1 |
| chr4_+_78978724 | 0.17 |
ENST00000325942.6
ENST00000264895.6 ENST00000264899.6 |
FRAS1
|
Fraser syndrome 1 |
| chr3_-_178969403 | 0.16 |
ENST00000314235.5
ENST00000392685.2 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr8_+_95653427 | 0.16 |
ENST00000454170.2
|
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr2_+_102928009 | 0.16 |
ENST00000404917.2
ENST00000447231.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr19_+_41305085 | 0.16 |
ENST00000303961.4
|
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr6_-_36953833 | 0.16 |
ENST00000538808.1
ENST00000460219.1 ENST00000373616.5 ENST00000373627.5 |
MTCH1
|
mitochondrial carrier 1 |
| chr7_+_7606605 | 0.16 |
ENST00000456533.1
|
MIOS
|
missing oocyte, meiosis regulator, homolog (Drosophila) |
| chr4_-_23735183 | 0.16 |
ENST00000507666.1
|
RP11-380P13.2
|
RP11-380P13.2 |
| chr6_-_117086873 | 0.16 |
ENST00000368557.4
|
FAM162B
|
family with sequence similarity 162, member B |
| chr16_+_28834303 | 0.16 |
ENST00000340394.8
ENST00000325215.6 ENST00000395547.2 ENST00000336783.4 ENST00000382686.4 ENST00000564304.1 |
ATXN2L
|
ataxin 2-like |
| chr8_+_9183618 | 0.16 |
ENST00000518619.1
|
RP11-115J16.1
|
RP11-115J16.1 |
| chr19_-_16045619 | 0.15 |
ENST00000402119.4
|
CYP4F11
|
cytochrome P450, family 4, subfamily F, polypeptide 11 |
| chr19_+_44037334 | 0.15 |
ENST00000314228.5
|
ZNF575
|
zinc finger protein 575 |
| chr5_+_175488258 | 0.15 |
ENST00000512862.1
|
FAM153B
|
family with sequence similarity 153, member B |
| chr9_-_139839064 | 0.15 |
ENST00000325285.3
ENST00000428398.1 |
FBXW5
|
F-box and WD repeat domain containing 5 |
| chr11_-_65667997 | 0.15 |
ENST00000312562.2
ENST00000534222.1 |
FOSL1
|
FOS-like antigen 1 |
| chrX_+_23928500 | 0.15 |
ENST00000435707.1
|
CXorf58
|
chromosome X open reading frame 58 |
| chr1_-_115053781 | 0.15 |
ENST00000358465.2
ENST00000369543.2 |
TRIM33
|
tripartite motif containing 33 |
| chr11_-_107729504 | 0.14 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr2_+_198557830 | 0.14 |
ENST00000409845.1
|
AC011997.1
|
Uncharacterized protein |
| chr2_+_235903480 | 0.14 |
ENST00000454947.1
|
SH3BP4
|
SH3-domain binding protein 4 |
| chr2_-_232329186 | 0.14 |
ENST00000322723.4
|
NCL
|
nucleolin |
| chrX_-_13835147 | 0.14 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr2_+_220363579 | 0.14 |
ENST00000313597.5
ENST00000373917.3 ENST00000358215.3 ENST00000373908.1 ENST00000455657.1 ENST00000435316.1 ENST00000341142.3 |
GMPPA
|
GDP-mannose pyrophosphorylase A |
| chr22_+_29664305 | 0.14 |
ENST00000414183.2
ENST00000333395.6 ENST00000455726.1 ENST00000332035.6 |
EWSR1
|
EWS RNA-binding protein 1 |
| chr5_-_39270725 | 0.14 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr19_+_50270219 | 0.13 |
ENST00000354293.5
ENST00000359032.5 |
AP2A1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr17_-_34591208 | 0.13 |
ENST00000336331.5
|
TBC1D3C
|
TBC1 domain family, member 3C |
| chr9_+_138392483 | 0.13 |
ENST00000241600.5
|
MRPS2
|
mitochondrial ribosomal protein S2 |
| chr14_+_94640633 | 0.13 |
ENST00000304338.3
|
PPP4R4
|
protein phosphatase 4, regulatory subunit 4 |
| chr19_+_7599597 | 0.13 |
ENST00000414982.3
ENST00000450331.3 |
PNPLA6
|
patatin-like phospholipase domain containing 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of VDR
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 0.1 | 0.4 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.7 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.1 | 0.9 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.1 | 0.3 | GO:1904717 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.1 | 0.3 | GO:0015920 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) lipopolysaccharide transport(GO:0015920) |
| 0.1 | 0.4 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.1 | 0.4 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.1 | 0.3 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.1 | 0.3 | GO:0043311 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.1 | 0.3 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 1.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.9 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 0.9 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 0.5 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.7 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 0.2 | GO:0006788 | heme oxidation(GO:0006788) negative regulation of mast cell cytokine production(GO:0032764) regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.1 | 0.6 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 | 0.2 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.1 | 0.6 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.2 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.1 | 0.3 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.1 | 0.3 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.7 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.3 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.3 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.3 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.4 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.3 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 1.0 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.6 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 1.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.0 | 0.9 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.3 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.4 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.2 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 0.2 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.3 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.3 | GO:0051873 | disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.0 | 0.1 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.0 | 0.2 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.3 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.3 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.0 | 0.2 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.2 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.4 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.5 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.0 | 0.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.5 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.2 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.3 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.2 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.4 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) |
| 0.0 | 0.3 | GO:0045324 | late endosome to vacuole transport(GO:0045324) ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.5 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.0 | GO:0000494 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.0 | 0.2 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.0 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.4 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.1 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.8 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.4 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.0 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.0 | 1.4 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.6 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 0.2 | GO:0036149 | layer formation in cerebral cortex(GO:0021819) phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.3 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.0 | 0.1 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.1 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 0.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.5 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 1.7 | GO:0090630 | activation of GTPase activity(GO:0090630) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.7 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 0.4 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 0.3 | GO:0036029 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.1 | 0.9 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.4 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.1 | 0.3 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.9 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.6 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.2 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.6 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.3 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.4 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.3 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 1.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.5 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.8 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.1 | 0.3 | GO:0045485 | omega-6 fatty acid desaturase activity(GO:0045485) |
| 0.1 | 0.9 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 0.3 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.1 | 0.6 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.1 | 0.3 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.1 | 0.3 | GO:0004397 | histidine ammonia-lyase activity(GO:0004397) |
| 0.1 | 0.3 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 0.4 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.1 | 0.5 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 0.3 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 0.4 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.2 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.3 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 0.6 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 0.2 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.2 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.5 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.8 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.3 | GO:0070891 | lipoteichoic acid binding(GO:0070891) lipopeptide binding(GO:0071723) |
| 0.0 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 1.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.1 | GO:0070362 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.0 | 0.3 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.6 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 1.0 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.9 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.0 | 0.7 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.2 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.2 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.0 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.2 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 1.5 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.9 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.5 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.4 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.6 | ST GAQ PATHWAY | G alpha q Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.8 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.7 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.2 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.0 | 0.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.1 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.6 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 1.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.7 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.4 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |