Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
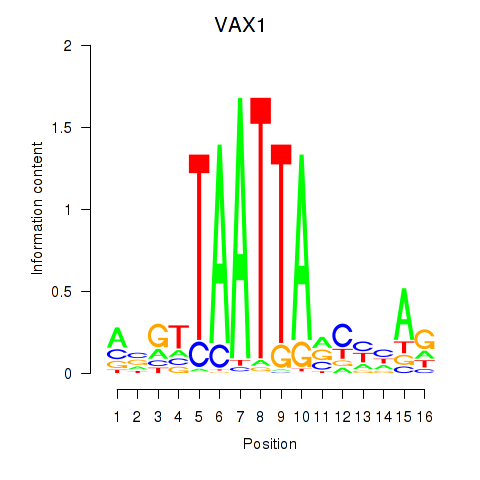
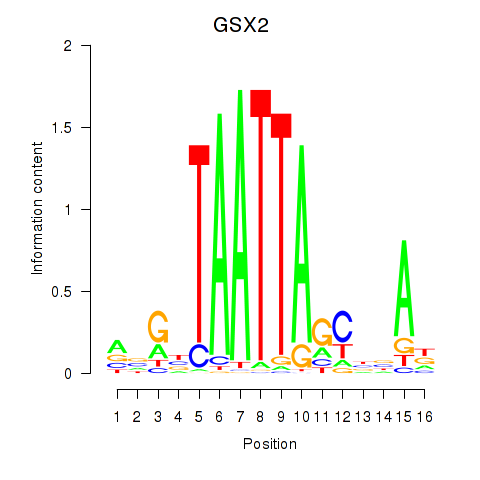
Results for VAX1_GSX2
Z-value: 0.51
Transcription factors associated with VAX1_GSX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
VAX1
|
ENSG00000148704.8 | ventral anterior homeobox 1 |
|
GSX2
|
ENSG00000180613.6 | GS homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| VAX1 | hg19_v2_chr10_-_118897806_118897817 | 0.58 | 4.2e-01 | Click! |
Activity profile of VAX1_GSX2 motif
Sorted Z-values of VAX1_GSX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_66536248 | 0.77 |
ENST00000514260.1
ENST00000507117.1 |
RP11-807H7.1
|
RP11-807H7.1 |
| chr15_-_37393406 | 0.46 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr4_-_89442940 | 0.46 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chr16_+_53133070 | 0.38 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr19_+_9203855 | 0.37 |
ENST00000429566.3
|
OR1M1
|
olfactory receptor, family 1, subfamily M, member 1 |
| chr14_+_20187174 | 0.30 |
ENST00000557414.1
|
OR4N2
|
olfactory receptor, family 4, subfamily N, member 2 |
| chr7_-_99716914 | 0.30 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr17_-_57229155 | 0.27 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr17_-_64225508 | 0.26 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr7_-_35013217 | 0.26 |
ENST00000446375.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr1_-_197115818 | 0.25 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr4_-_25865159 | 0.23 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr13_-_41593425 | 0.23 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr13_-_46626820 | 0.22 |
ENST00000428921.1
|
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr2_+_44502597 | 0.21 |
ENST00000260649.6
ENST00000409387.1 |
SLC3A1
|
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr11_-_129062093 | 0.21 |
ENST00000310343.9
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr3_+_149191723 | 0.20 |
ENST00000305354.4
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr1_+_28261492 | 0.19 |
ENST00000373894.3
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr2_-_188419078 | 0.19 |
ENST00000437725.1
ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr16_+_12059091 | 0.19 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr1_-_202897724 | 0.19 |
ENST00000435533.3
ENST00000367258.1 |
KLHL12
|
kelch-like family member 12 |
| chr13_+_111748183 | 0.18 |
ENST00000422994.1
|
LINC00368
|
long intergenic non-protein coding RNA 368 |
| chr10_+_91461337 | 0.17 |
ENST00000260753.4
ENST00000416354.1 ENST00000394289.2 ENST00000371728.3 |
KIF20B
|
kinesin family member 20B |
| chr1_-_109618566 | 0.17 |
ENST00000338366.5
|
TAF13
|
TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa |
| chr15_+_96904487 | 0.17 |
ENST00000600790.1
|
AC087477.1
|
Uncharacterized protein |
| chr12_+_28410128 | 0.17 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr1_-_207226313 | 0.16 |
ENST00000367084.1
|
YOD1
|
YOD1 deubiquitinase |
| chr9_-_5339873 | 0.16 |
ENST00000223862.1
ENST00000223858.4 |
RLN1
|
relaxin 1 |
| chr17_+_73539339 | 0.16 |
ENST00000581713.1
|
LLGL2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr12_-_15815626 | 0.15 |
ENST00000540613.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr4_+_95128748 | 0.15 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr6_-_160209471 | 0.15 |
ENST00000539948.1
|
TCP1
|
t-complex 1 |
| chr16_+_72088376 | 0.15 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr17_-_39341594 | 0.15 |
ENST00000398472.1
|
KRTAP4-1
|
keratin associated protein 4-1 |
| chr12_-_10978957 | 0.15 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr3_+_138340067 | 0.15 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr6_-_108278456 | 0.15 |
ENST00000429168.1
|
SEC63
|
SEC63 homolog (S. cerevisiae) |
| chr2_-_44588624 | 0.15 |
ENST00000438314.1
ENST00000409936.1 |
PREPL
|
prolyl endopeptidase-like |
| chr21_-_35899113 | 0.15 |
ENST00000492600.1
ENST00000481448.1 ENST00000381132.2 |
RCAN1
|
regulator of calcineurin 1 |
| chr11_-_795170 | 0.15 |
ENST00000481290.1
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr17_-_45266542 | 0.15 |
ENST00000531206.1
ENST00000527547.1 ENST00000446365.2 ENST00000575483.1 ENST00000066544.3 |
CDC27
|
cell division cycle 27 |
| chr13_+_36050881 | 0.14 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
| chr11_+_100862811 | 0.14 |
ENST00000303130.2
|
TMEM133
|
transmembrane protein 133 |
| chr9_-_5304432 | 0.14 |
ENST00000416837.1
ENST00000308420.3 |
RLN2
|
relaxin 2 |
| chr18_+_46065393 | 0.14 |
ENST00000256413.3
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chrX_+_43515467 | 0.14 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr21_-_39705323 | 0.14 |
ENST00000436845.1
|
AP001422.3
|
AP001422.3 |
| chr14_+_67831576 | 0.14 |
ENST00000555876.1
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
| chr6_+_130339710 | 0.14 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr3_+_158519654 | 0.14 |
ENST00000415822.2
ENST00000392813.4 ENST00000264266.8 |
MFSD1
|
major facilitator superfamily domain containing 1 |
| chr1_-_150669500 | 0.13 |
ENST00000271732.3
|
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr2_-_136678123 | 0.13 |
ENST00000422708.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr12_+_16500037 | 0.13 |
ENST00000536371.1
ENST00000010404.2 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr2_-_44588694 | 0.13 |
ENST00000409957.1
|
PREPL
|
prolyl endopeptidase-like |
| chr8_-_97273807 | 0.12 |
ENST00000517720.1
ENST00000287025.3 ENST00000523821.1 |
MTERFD1
|
MTERF domain containing 1 |
| chr12_-_64062583 | 0.12 |
ENST00000542209.1
|
DPY19L2
|
dpy-19-like 2 (C. elegans) |
| chr2_-_44588679 | 0.12 |
ENST00000409411.1
|
PREPL
|
prolyl endopeptidase-like |
| chr6_-_116833500 | 0.12 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr1_-_68698197 | 0.12 |
ENST00000370973.2
ENST00000370971.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr2_+_44502630 | 0.11 |
ENST00000410056.3
ENST00000409741.1 ENST00000409229.3 |
SLC3A1
|
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr9_-_5830768 | 0.11 |
ENST00000381506.3
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr6_-_82957433 | 0.11 |
ENST00000306270.7
|
IBTK
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr15_+_36871983 | 0.11 |
ENST00000437989.2
ENST00000569302.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr1_+_81771806 | 0.11 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chr13_+_40229764 | 0.11 |
ENST00000416691.1
ENST00000422759.2 ENST00000455146.3 |
COG6
|
component of oligomeric golgi complex 6 |
| chr11_-_33774944 | 0.10 |
ENST00000532057.1
ENST00000531080.1 |
FBXO3
|
F-box protein 3 |
| chr15_-_65903407 | 0.10 |
ENST00000395644.4
ENST00000567744.1 ENST00000568573.1 ENST00000562830.1 ENST00000569491.1 ENST00000561769.1 |
VWA9
|
von Willebrand factor A domain containing 9 |
| chr14_+_57671888 | 0.10 |
ENST00000391612.1
|
AL391152.1
|
AL391152.1 |
| chr6_+_26440700 | 0.09 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr9_+_125133315 | 0.09 |
ENST00000223423.4
ENST00000362012.2 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr7_+_77428066 | 0.09 |
ENST00000422959.2
ENST00000307305.8 ENST00000424760.1 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr13_-_46626847 | 0.09 |
ENST00000242848.4
ENST00000282007.3 |
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr10_-_92681033 | 0.09 |
ENST00000371697.3
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr11_+_115498761 | 0.09 |
ENST00000424313.2
|
AP000997.1
|
AP000997.1 |
| chr17_-_7167279 | 0.09 |
ENST00000571932.2
|
CLDN7
|
claudin 7 |
| chr11_-_27722021 | 0.09 |
ENST00000356660.4
ENST00000418212.1 ENST00000533246.1 |
BDNF
|
brain-derived neurotrophic factor |
| chr7_-_76955563 | 0.08 |
ENST00000441833.2
|
GSAP
|
gamma-secretase activating protein |
| chr6_-_38670897 | 0.08 |
ENST00000373365.4
|
GLO1
|
glyoxalase I |
| chr4_-_109541539 | 0.08 |
ENST00000509984.1
ENST00000507248.1 ENST00000506795.1 |
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr4_-_103749105 | 0.08 |
ENST00000394801.4
ENST00000394804.2 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr8_+_75262612 | 0.08 |
ENST00000220822.7
|
GDAP1
|
ganglioside induced differentiation associated protein 1 |
| chr1_-_77685084 | 0.08 |
ENST00000370812.3
ENST00000359130.1 ENST00000445065.1 ENST00000370813.5 |
PIGK
|
phosphatidylinositol glycan anchor biosynthesis, class K |
| chr19_-_52307357 | 0.08 |
ENST00000594900.1
|
FPR1
|
formyl peptide receptor 1 |
| chr3_+_44840679 | 0.08 |
ENST00000425755.1
|
KIF15
|
kinesin family member 15 |
| chrX_+_107334895 | 0.08 |
ENST00000372232.3
ENST00000345734.3 ENST00000372254.3 |
ATG4A
|
autophagy related 4A, cysteine peptidase |
| chrX_+_108779004 | 0.08 |
ENST00000218004.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr2_+_198380289 | 0.08 |
ENST00000233892.4
ENST00000409916.1 |
MOB4
|
MOB family member 4, phocein |
| chr11_-_96076334 | 0.08 |
ENST00000524717.1
|
MAML2
|
mastermind-like 2 (Drosophila) |
| chr15_-_75748143 | 0.08 |
ENST00000568431.1
ENST00000568309.1 ENST00000568190.1 ENST00000570115.1 ENST00000564778.1 |
SIN3A
|
SIN3 transcription regulator family member A |
| chr2_-_3521518 | 0.08 |
ENST00000382093.5
|
ADI1
|
acireductone dioxygenase 1 |
| chr10_-_27529486 | 0.08 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr5_-_98262240 | 0.08 |
ENST00000284049.3
|
CHD1
|
chromodomain helicase DNA binding protein 1 |
| chr16_+_29832634 | 0.08 |
ENST00000565164.1
ENST00000570234.1 |
MVP
|
major vault protein |
| chr8_+_27238147 | 0.07 |
ENST00000412793.1
|
PTK2B
|
protein tyrosine kinase 2 beta |
| chr4_-_164534657 | 0.07 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr17_+_28443819 | 0.07 |
ENST00000479218.2
|
NSRP1
|
nuclear speckle splicing regulatory protein 1 |
| chr16_-_66764119 | 0.07 |
ENST00000569320.1
|
DYNC1LI2
|
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr4_-_103749313 | 0.07 |
ENST00000394803.5
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr1_+_162336686 | 0.07 |
ENST00000420220.1
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr5_-_94890648 | 0.07 |
ENST00000513823.1
ENST00000514952.1 ENST00000358746.2 |
TTC37
|
tetratricopeptide repeat domain 37 |
| chr22_-_18923655 | 0.07 |
ENST00000438924.1
ENST00000457083.1 ENST00000420436.1 ENST00000334029.2 ENST00000357068.6 |
PRODH
|
proline dehydrogenase (oxidase) 1 |
| chr11_-_111649015 | 0.07 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr12_+_16500599 | 0.07 |
ENST00000535309.1
ENST00000540056.1 ENST00000396209.1 ENST00000540126.1 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr10_+_99205894 | 0.07 |
ENST00000370854.3
ENST00000393760.1 ENST00000414567.1 ENST00000370846.4 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chr6_+_28317685 | 0.07 |
ENST00000252211.2
ENST00000341464.5 ENST00000377255.3 |
ZKSCAN3
|
zinc finger with KRAB and SCAN domains 3 |
| chr22_-_32651326 | 0.06 |
ENST00000266086.4
|
SLC5A4
|
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr17_-_64187973 | 0.06 |
ENST00000583358.1
ENST00000392769.2 |
CEP112
|
centrosomal protein 112kDa |
| chr5_-_145562147 | 0.06 |
ENST00000545646.1
ENST00000274562.9 ENST00000510191.1 ENST00000394434.2 |
LARS
|
leucyl-tRNA synthetase |
| chr3_+_130569429 | 0.06 |
ENST00000505330.1
ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr20_+_43990576 | 0.06 |
ENST00000372727.1
ENST00000414310.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr8_-_102803163 | 0.06 |
ENST00000523645.1
ENST00000520346.1 ENST00000220931.6 ENST00000522448.1 ENST00000522951.1 ENST00000522252.1 ENST00000519098.1 |
NCALD
|
neurocalcin delta |
| chr1_+_236958554 | 0.06 |
ENST00000366577.5
ENST00000418145.2 |
MTR
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr7_+_37723450 | 0.06 |
ENST00000447769.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr7_+_156433573 | 0.06 |
ENST00000311822.8
|
RNF32
|
ring finger protein 32 |
| chr14_-_90085458 | 0.06 |
ENST00000345097.4
ENST00000555855.1 ENST00000555353.1 |
FOXN3
|
forkhead box N3 |
| chr15_-_75748115 | 0.06 |
ENST00000360439.4
|
SIN3A
|
SIN3 transcription regulator family member A |
| chr13_+_78315295 | 0.06 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr11_+_45376922 | 0.06 |
ENST00000524410.1
ENST00000524488.1 ENST00000524565.1 |
RP11-430H10.1
|
RP11-430H10.1 |
| chr2_-_101925055 | 0.06 |
ENST00000295317.3
|
RNF149
|
ring finger protein 149 |
| chr3_+_62304648 | 0.06 |
ENST00000462069.1
ENST00000232519.5 ENST00000465142.1 |
C3orf14
|
chromosome 3 open reading frame 14 |
| chr8_+_9413410 | 0.06 |
ENST00000520408.1
ENST00000310430.6 ENST00000522110.1 |
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr2_+_187371440 | 0.06 |
ENST00000445547.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr12_+_20963632 | 0.06 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr14_+_105864847 | 0.06 |
ENST00000451127.2
|
TEX22
|
testis expressed 22 |
| chr4_-_169239921 | 0.05 |
ENST00000514995.1
ENST00000393743.3 |
DDX60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr6_+_26365387 | 0.05 |
ENST00000532865.1
ENST00000530653.1 ENST00000527417.1 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr12_-_48744554 | 0.05 |
ENST00000544117.2
ENST00000548932.1 ENST00000549125.1 ENST00000301042.3 ENST00000547026.1 |
ZNF641
|
zinc finger protein 641 |
| chr8_+_105602961 | 0.05 |
ENST00000521923.1
|
RP11-127H5.1
|
Uncharacterized protein |
| chr4_-_103748880 | 0.05 |
ENST00000453744.2
ENST00000349311.8 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr7_+_99202003 | 0.05 |
ENST00000609449.1
|
GS1-259H13.2
|
GS1-259H13.2 |
| chr7_-_111424506 | 0.05 |
ENST00000450156.1
ENST00000494651.2 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr8_-_55014018 | 0.05 |
ENST00000521352.1
|
LYPLA1
|
lysophospholipase I |
| chr15_+_75491203 | 0.05 |
ENST00000562637.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr1_+_186369704 | 0.05 |
ENST00000574641.1
|
OCLM
|
oculomedin |
| chrX_+_107334983 | 0.05 |
ENST00000457035.1
ENST00000545696.1 |
ATG4A
|
autophagy related 4A, cysteine peptidase |
| chr12_+_64798095 | 0.05 |
ENST00000332707.5
|
XPOT
|
exportin, tRNA |
| chr15_+_58702742 | 0.05 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr7_-_111424462 | 0.05 |
ENST00000437129.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr3_-_105587879 | 0.05 |
ENST00000264122.4
ENST00000403724.1 ENST00000405772.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr12_+_16500571 | 0.05 |
ENST00000543076.1
ENST00000396210.3 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr14_+_104182105 | 0.05 |
ENST00000311141.2
|
ZFYVE21
|
zinc finger, FYVE domain containing 21 |
| chr3_+_101504200 | 0.05 |
ENST00000422132.1
|
NXPE3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr14_+_39583427 | 0.05 |
ENST00000308317.6
ENST00000396249.2 ENST00000250379.8 ENST00000534684.2 ENST00000527381.1 |
GEMIN2
|
gem (nuclear organelle) associated protein 2 |
| chr17_+_28443799 | 0.05 |
ENST00000584423.1
ENST00000247026.5 |
NSRP1
|
nuclear speckle splicing regulatory protein 1 |
| chr22_-_36903069 | 0.05 |
ENST00000216187.6
ENST00000423980.1 |
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr10_+_99205959 | 0.05 |
ENST00000352634.4
ENST00000353979.3 ENST00000370842.2 ENST00000345745.5 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chr8_+_107738240 | 0.05 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr11_+_66276550 | 0.05 |
ENST00000419755.3
|
CTD-3074O7.11
|
Bardet-Biedl syndrome 1 protein |
| chr2_+_32288657 | 0.05 |
ENST00000345662.1
|
SPAST
|
spastin |
| chrX_+_37639264 | 0.05 |
ENST00000378588.4
|
CYBB
|
cytochrome b-245, beta polypeptide |
| chr2_+_102953608 | 0.05 |
ENST00000311734.2
ENST00000409584.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr12_+_20963647 | 0.05 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr20_+_816695 | 0.05 |
ENST00000246100.3
|
FAM110A
|
family with sequence similarity 110, member A |
| chr14_-_70883708 | 0.05 |
ENST00000256366.4
|
SYNJ2BP
|
synaptojanin 2 binding protein |
| chr4_+_80584903 | 0.04 |
ENST00000506460.1
|
RP11-452C8.1
|
RP11-452C8.1 |
| chrX_-_16887963 | 0.04 |
ENST00000380084.4
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr2_+_32288725 | 0.04 |
ENST00000315285.3
|
SPAST
|
spastin |
| chr14_+_101359265 | 0.04 |
ENST00000599197.1
|
AL117190.3
|
Esophagus cancer-related gene-2 interaction susceptibility protein; Uncharacterized protein |
| chr8_+_107738343 | 0.04 |
ENST00000521592.1
|
OXR1
|
oxidation resistance 1 |
| chr13_+_73632897 | 0.04 |
ENST00000377687.4
|
KLF5
|
Kruppel-like factor 5 (intestinal) |
| chr15_+_89631381 | 0.04 |
ENST00000352732.5
|
ABHD2
|
abhydrolase domain containing 2 |
| chr11_+_101918153 | 0.04 |
ENST00000434758.2
ENST00000526781.1 ENST00000534360.1 |
C11orf70
|
chromosome 11 open reading frame 70 |
| chr7_+_77428149 | 0.04 |
ENST00000415251.2
ENST00000275575.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr1_-_54411255 | 0.04 |
ENST00000371377.3
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr18_-_46895066 | 0.04 |
ENST00000583225.1
ENST00000584983.1 ENST00000583280.1 ENST00000581738.1 |
DYM
|
dymeclin |
| chr17_+_18086392 | 0.04 |
ENST00000541285.1
|
ALKBH5
|
alkB, alkylation repair homolog 5 (E. coli) |
| chr3_+_51851612 | 0.04 |
ENST00000456080.1
|
IQCF3
|
IQ motif containing F3 |
| chr17_-_4938712 | 0.04 |
ENST00000254853.5
ENST00000424747.1 |
SLC52A1
|
solute carrier family 52 (riboflavin transporter), member 1 |
| chr4_+_119809984 | 0.04 |
ENST00000307142.4
ENST00000448416.2 ENST00000429713.2 |
SYNPO2
|
synaptopodin 2 |
| chr5_+_66300446 | 0.04 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr2_+_11682790 | 0.04 |
ENST00000389825.3
ENST00000381483.2 |
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr9_+_131062367 | 0.04 |
ENST00000601297.1
|
AL359091.2
|
CDNA: FLJ21673 fis, clone COL09042; HCG2036511; Uncharacterized protein |
| chr9_+_90112767 | 0.04 |
ENST00000408954.3
|
DAPK1
|
death-associated protein kinase 1 |
| chr18_-_54305658 | 0.04 |
ENST00000586262.1
ENST00000217515.6 |
TXNL1
|
thioredoxin-like 1 |
| chr7_+_99717230 | 0.04 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr1_-_68698222 | 0.04 |
ENST00000370976.3
ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr3_-_196911002 | 0.04 |
ENST00000452595.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr4_+_48492269 | 0.04 |
ENST00000327939.4
|
ZAR1
|
zygote arrest 1 |
| chr1_-_151762900 | 0.04 |
ENST00000440583.2
|
TDRKH
|
tudor and KH domain containing |
| chr1_+_207226574 | 0.04 |
ENST00000367080.3
ENST00000367079.2 |
PFKFB2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr7_-_14029283 | 0.03 |
ENST00000433547.1
ENST00000405192.2 |
ETV1
|
ets variant 1 |
| chr16_+_31885079 | 0.03 |
ENST00000300870.10
ENST00000394846.3 |
ZNF267
|
zinc finger protein 267 |
| chr5_-_143550159 | 0.03 |
ENST00000448443.2
ENST00000513112.1 ENST00000519064.1 ENST00000274496.5 |
YIPF5
|
Yip1 domain family, member 5 |
| chr17_+_48823975 | 0.03 |
ENST00000513969.1
ENST00000503728.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr6_+_110501621 | 0.03 |
ENST00000368930.1
ENST00000307731.1 |
CDC40
|
cell division cycle 40 |
| chr12_-_10282742 | 0.03 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr22_-_39637135 | 0.03 |
ENST00000440375.1
|
PDGFB
|
platelet-derived growth factor beta polypeptide |
| chr2_-_208031943 | 0.03 |
ENST00000421199.1
ENST00000457962.1 |
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr15_+_90118723 | 0.03 |
ENST00000560985.1
|
TICRR
|
TOPBP1-interacting checkpoint and replication regulator |
| chr3_+_111630451 | 0.03 |
ENST00000495180.1
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr11_-_795286 | 0.03 |
ENST00000533385.1
ENST00000527723.1 |
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr4_+_41614909 | 0.03 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr17_-_19364269 | 0.03 |
ENST00000421796.2
ENST00000585389.1 ENST00000609249.1 |
AC004448.5
|
AC004448.5 |
| chr15_-_55562451 | 0.03 |
ENST00000568803.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr1_+_84630645 | 0.03 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr22_-_36903101 | 0.03 |
ENST00000397224.4
|
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr11_-_795400 | 0.03 |
ENST00000526152.1
ENST00000456706.2 ENST00000528936.1 |
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr6_+_26365443 | 0.03 |
ENST00000527422.1
ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr14_+_104182061 | 0.03 |
ENST00000216602.6
|
ZFYVE21
|
zinc finger, FYVE domain containing 21 |
| chr17_+_74846230 | 0.03 |
ENST00000592919.1
|
LINC00868
|
long intergenic non-protein coding RNA 868 |
Network of associatons between targets according to the STRING database.
First level regulatory network of VAX1_GSX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.2 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.1 | 0.2 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.0 | 0.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.3 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.2 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.2 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.2 | GO:0051836 | translocation of peptides or proteins into host(GO:0042000) translocation of peptides or proteins into host cell cytoplasm(GO:0044053) translocation of molecules into host(GO:0044417) translocation of peptides or proteins into other organism involved in symbiotic interaction(GO:0051808) translocation of molecules into other organism involved in symbiotic interaction(GO:0051836) |
| 0.0 | 0.1 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.4 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.2 | GO:1903490 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.0 | 0.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.2 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.3 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:0006429 | glutaminyl-tRNA aminoacylation(GO:0006425) leucyl-tRNA aminoacylation(GO:0006429) |
| 0.0 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.7 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.0 | 0.1 | GO:2000537 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.1 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) astrocyte end-foot(GO:0097450) |
| 0.0 | 0.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.5 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.1 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.3 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.2 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0004823 | glutamine-tRNA ligase activity(GO:0004819) leucine-tRNA ligase activity(GO:0004823) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.2 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 0.0 | 0.0 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.7 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.4 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.4 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.2 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |