Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
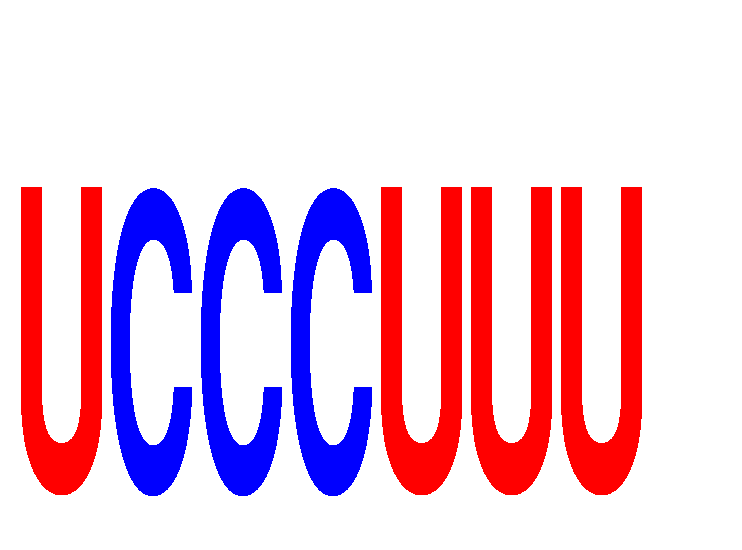
Results for UCCCUUU
Z-value: 0.69
miRNA associated with seed UCCCUUU
| Name | miRBASE accession |
|---|---|
|
hsa-miR-204-5p
|
MIMAT0000265 |
|
hsa-miR-211-5p
|
MIMAT0000268 |
Activity profile of UCCCUUU motif
Sorted Z-values of UCCCUUU motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_98605902 | 0.42 |
ENST00000460070.1
ENST00000481455.1 ENST00000261574.5 ENST00000493281.1 ENST00000463157.1 ENST00000471898.1 ENST00000489058.1 ENST00000481689.1 |
IPO5
|
importin 5 |
| chr12_+_20522179 | 0.41 |
ENST00000359062.3
|
PDE3A
|
phosphodiesterase 3A, cGMP-inhibited |
| chr5_-_693500 | 0.41 |
ENST00000360578.5
|
TPPP
|
tubulin polymerization promoting protein |
| chr1_+_65613217 | 0.40 |
ENST00000545314.1
|
AK4
|
adenylate kinase 4 |
| chr5_-_142783175 | 0.36 |
ENST00000231509.3
ENST00000394464.2 |
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr8_-_91658303 | 0.31 |
ENST00000458549.2
|
TMEM64
|
transmembrane protein 64 |
| chr20_+_56884752 | 0.29 |
ENST00000244040.3
|
RAB22A
|
RAB22A, member RAS oncogene family |
| chr11_-_75062730 | 0.29 |
ENST00000420843.2
ENST00000360025.3 |
ARRB1
|
arrestin, beta 1 |
| chr15_+_57210818 | 0.29 |
ENST00000438423.2
ENST00000267811.5 ENST00000452095.2 ENST00000559609.1 ENST00000333725.5 |
TCF12
|
transcription factor 12 |
| chr1_-_70671216 | 0.29 |
ENST00000370952.3
|
LRRC40
|
leucine rich repeat containing 40 |
| chr6_-_159239257 | 0.28 |
ENST00000337147.7
ENST00000392177.4 |
EZR
|
ezrin |
| chr10_+_69644404 | 0.28 |
ENST00000212015.6
|
SIRT1
|
sirtuin 1 |
| chr10_-_81205373 | 0.27 |
ENST00000372336.3
|
ZCCHC24
|
zinc finger, CCHC domain containing 24 |
| chr1_+_154377669 | 0.27 |
ENST00000368485.3
ENST00000344086.4 |
IL6R
|
interleukin 6 receptor |
| chr17_+_55162453 | 0.26 |
ENST00000575322.1
ENST00000337714.3 ENST00000314126.3 |
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr6_+_1610681 | 0.26 |
ENST00000380874.2
|
FOXC1
|
forkhead box C1 |
| chr3_+_30648066 | 0.25 |
ENST00000359013.4
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa) |
| chrX_-_131352152 | 0.24 |
ENST00000342983.2
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr6_+_147830063 | 0.23 |
ENST00000367474.1
|
SAMD5
|
sterile alpha motif domain containing 5 |
| chr11_-_74204742 | 0.22 |
ENST00000310109.4
|
LIPT2
|
lipoyl(octanoyl) transferase 2 (putative) |
| chrX_-_135056216 | 0.21 |
ENST00000305963.2
|
MMGT1
|
membrane magnesium transporter 1 |
| chrX_-_20284958 | 0.20 |
ENST00000379565.3
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr4_-_114682936 | 0.20 |
ENST00000454265.2
ENST00000429180.1 ENST00000418639.2 ENST00000394526.2 ENST00000296402.5 |
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta |
| chr17_+_30469473 | 0.19 |
ENST00000333942.6
ENST00000358365.3 ENST00000583994.1 ENST00000545287.2 |
RHOT1
|
ras homolog family member T1 |
| chr16_-_30798492 | 0.18 |
ENST00000262525.4
|
ZNF629
|
zinc finger protein 629 |
| chr2_-_157189180 | 0.18 |
ENST00000539077.1
ENST00000424077.1 ENST00000426264.1 ENST00000339562.4 ENST00000421709.1 |
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr11_-_57283159 | 0.18 |
ENST00000533263.1
ENST00000278426.3 |
SLC43A1
|
solute carrier family 43 (amino acid system L transporter), member 1 |
| chr3_-_142166904 | 0.17 |
ENST00000264951.4
|
XRN1
|
5'-3' exoribonuclease 1 |
| chr18_+_9136758 | 0.16 |
ENST00000383440.2
ENST00000262126.4 ENST00000577992.1 |
ANKRD12
|
ankyrin repeat domain 12 |
| chr4_-_23891693 | 0.16 |
ENST00000264867.2
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr7_+_138145076 | 0.16 |
ENST00000343526.4
|
TRIM24
|
tripartite motif containing 24 |
| chr6_-_39197226 | 0.16 |
ENST00000359534.3
|
KCNK5
|
potassium channel, subfamily K, member 5 |
| chr19_+_34919257 | 0.16 |
ENST00000246548.4
ENST00000590048.2 |
UBA2
|
ubiquitin-like modifier activating enzyme 2 |
| chr16_+_46918235 | 0.15 |
ENST00000340124.4
|
GPT2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr1_-_39325431 | 0.15 |
ENST00000373001.3
|
RRAGC
|
Ras-related GTP binding C |
| chr11_-_27494279 | 0.15 |
ENST00000379214.4
|
LGR4
|
leucine-rich repeat containing G protein-coupled receptor 4 |
| chrX_-_15872914 | 0.14 |
ENST00000380291.1
ENST00000545766.1 ENST00000421527.2 ENST00000329235.2 |
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr5_+_95066823 | 0.14 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr19_-_49149553 | 0.14 |
ENST00000084798.4
|
CA11
|
carbonic anhydrase XI |
| chr6_-_8435706 | 0.14 |
ENST00000379660.4
|
SLC35B3
|
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B3 |
| chr19_-_4066890 | 0.13 |
ENST00000322357.4
|
ZBTB7A
|
zinc finger and BTB domain containing 7A |
| chr13_-_31736027 | 0.13 |
ENST00000380406.5
ENST00000320027.5 ENST00000380405.4 |
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr3_-_15374033 | 0.13 |
ENST00000253688.5
ENST00000383791.3 |
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated) |
| chr12_+_4430371 | 0.13 |
ENST00000179259.4
|
C12orf5
|
chromosome 12 open reading frame 5 |
| chr7_+_142552792 | 0.13 |
ENST00000392957.2
ENST00000442129.1 |
EPHB6
|
EPH receptor B6 |
| chr10_+_126490354 | 0.13 |
ENST00000298492.5
|
FAM175B
|
family with sequence similarity 175, member B |
| chr11_+_34073195 | 0.13 |
ENST00000341394.4
|
CAPRIN1
|
cell cycle associated protein 1 |
| chr12_-_39299406 | 0.13 |
ENST00000331366.5
|
CPNE8
|
copine VIII |
| chr3_-_138553594 | 0.12 |
ENST00000477593.1
ENST00000483968.1 |
PIK3CB
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
| chr1_+_78470530 | 0.12 |
ENST00000370763.5
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chr2_-_152684977 | 0.12 |
ENST00000428992.2
ENST00000295087.8 |
ARL5A
|
ADP-ribosylation factor-like 5A |
| chr3_-_120068143 | 0.12 |
ENST00000295628.3
|
LRRC58
|
leucine rich repeat containing 58 |
| chr6_+_10556215 | 0.12 |
ENST00000316170.3
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr21_-_15755446 | 0.12 |
ENST00000544452.1
ENST00000285667.3 |
HSPA13
|
heat shock protein 70kDa family, member 13 |
| chr5_-_180288248 | 0.11 |
ENST00000512132.1
ENST00000506439.1 ENST00000502412.1 ENST00000359141.6 |
ZFP62
|
ZFP62 zinc finger protein |
| chr1_+_21835858 | 0.11 |
ENST00000539907.1
ENST00000540617.1 ENST00000374840.3 |
ALPL
|
alkaline phosphatase, liver/bone/kidney |
| chr5_-_95297678 | 0.11 |
ENST00000237853.4
|
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr7_-_127225620 | 0.11 |
ENST00000321407.2
|
GCC1
|
GRIP and coiled-coil domain containing 1 |
| chr10_+_60144782 | 0.11 |
ENST00000487519.1
|
TFAM
|
transcription factor A, mitochondrial |
| chr7_+_94285637 | 0.10 |
ENST00000482108.1
ENST00000488574.1 |
PEG10
|
paternally expressed 10 |
| chr17_+_28705921 | 0.10 |
ENST00000225719.4
|
CPD
|
carboxypeptidase D |
| chr1_+_64239657 | 0.10 |
ENST00000371080.1
ENST00000371079.1 |
ROR1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr9_+_33817461 | 0.09 |
ENST00000263228.3
|
UBE2R2
|
ubiquitin-conjugating enzyme E2R 2 |
| chr14_+_57735614 | 0.09 |
ENST00000261558.3
|
AP5M1
|
adaptor-related protein complex 5, mu 1 subunit |
| chrX_-_14891150 | 0.09 |
ENST00000452869.1
ENST00000398334.1 ENST00000324138.3 |
FANCB
|
Fanconi anemia, complementation group B |
| chr2_-_85555385 | 0.09 |
ENST00000377386.3
|
TGOLN2
|
trans-golgi network protein 2 |
| chr1_+_38259540 | 0.09 |
ENST00000397631.3
|
MANEAL
|
mannosidase, endo-alpha-like |
| chr7_-_32529973 | 0.09 |
ENST00000410044.1
ENST00000409987.1 ENST00000409782.1 ENST00000450169.2 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr6_+_127439749 | 0.08 |
ENST00000356698.4
|
RSPO3
|
R-spondin 3 |
| chr4_+_99916765 | 0.08 |
ENST00000296411.6
|
METAP1
|
methionyl aminopeptidase 1 |
| chr5_+_96271141 | 0.08 |
ENST00000231368.5
|
LNPEP
|
leucyl/cystinyl aminopeptidase |
| chr9_-_27573392 | 0.08 |
ENST00000380003.3
|
C9orf72
|
chromosome 9 open reading frame 72 |
| chr16_+_67063036 | 0.08 |
ENST00000290858.6
ENST00000564034.1 |
CBFB
|
core-binding factor, beta subunit |
| chr6_-_8102714 | 0.08 |
ENST00000502429.1
ENST00000429723.2 ENST00000507463.1 ENST00000379715.5 |
EEF1E1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr4_-_68566832 | 0.08 |
ENST00000420827.2
ENST00000322244.5 |
UBA6
|
ubiquitin-like modifier activating enzyme 6 |
| chr11_+_63706444 | 0.08 |
ENST00000377793.4
ENST00000456907.2 ENST00000539656.1 |
NAA40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr7_+_145813453 | 0.08 |
ENST00000361727.3
|
CNTNAP2
|
contactin associated protein-like 2 |
| chr8_+_42752053 | 0.08 |
ENST00000307602.4
|
HOOK3
|
hook microtubule-tethering protein 3 |
| chr18_+_32558208 | 0.08 |
ENST00000436190.2
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr3_+_169940153 | 0.07 |
ENST00000295797.4
|
PRKCI
|
protein kinase C, iota |
| chr6_-_90062543 | 0.07 |
ENST00000435041.2
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1 |
| chr2_+_201676256 | 0.07 |
ENST00000452206.1
ENST00000410110.2 ENST00000409600.1 |
BZW1
|
basic leucine zipper and W2 domains 1 |
| chr13_+_50571155 | 0.07 |
ENST00000420995.2
ENST00000378182.3 ENST00000356017.4 ENST00000457662.2 |
TRIM13
|
tripartite motif containing 13 |
| chr4_-_140098339 | 0.07 |
ENST00000394235.2
|
ELF2
|
E74-like factor 2 (ets domain transcription factor) |
| chrX_+_70752917 | 0.07 |
ENST00000373719.3
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr10_+_93683519 | 0.07 |
ENST00000265990.6
|
BTAF1
|
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated, 170kDa |
| chr4_-_38858428 | 0.07 |
ENST00000436693.2
ENST00000508254.1 ENST00000514655.1 ENST00000506146.1 |
TLR6
TLR1
|
toll-like receptor 6 toll-like receptor 1 |
| chr19_+_10527449 | 0.07 |
ENST00000592685.1
ENST00000380702.2 |
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr17_+_7608511 | 0.07 |
ENST00000226091.2
|
EFNB3
|
ephrin-B3 |
| chr13_-_42535214 | 0.07 |
ENST00000379310.3
ENST00000281496.6 |
VWA8
|
von Willebrand factor A domain containing 8 |
| chr11_+_58346584 | 0.06 |
ENST00000316059.6
|
ZFP91
|
ZFP91 zinc finger protein |
| chr4_-_36246060 | 0.06 |
ENST00000303965.4
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr15_+_52121822 | 0.06 |
ENST00000558455.1
ENST00000308580.7 |
TMOD3
|
tropomodulin 3 (ubiquitous) |
| chr15_+_43477455 | 0.06 |
ENST00000300213.4
|
CCNDBP1
|
cyclin D-type binding-protein 1 |
| chr15_-_34628951 | 0.06 |
ENST00000397707.2
ENST00000560611.1 |
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr3_-_9811595 | 0.06 |
ENST00000256460.3
|
CAMK1
|
calcium/calmodulin-dependent protein kinase I |
| chr5_+_148960931 | 0.06 |
ENST00000333677.6
|
ARHGEF37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr3_-_172428959 | 0.06 |
ENST00000475381.1
ENST00000538775.1 ENST00000273512.3 ENST00000543711.1 |
NCEH1
|
neutral cholesterol ester hydrolase 1 |
| chr15_-_27018175 | 0.06 |
ENST00000311550.5
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr2_-_211090162 | 0.05 |
ENST00000233710.3
|
ACADL
|
acyl-CoA dehydrogenase, long chain |
| chr12_+_62860581 | 0.05 |
ENST00000393632.2
ENST00000393630.3 ENST00000280379.6 ENST00000546600.1 ENST00000552738.1 ENST00000393629.2 ENST00000552115.1 |
MON2
|
MON2 homolog (S. cerevisiae) |
| chr10_+_18948311 | 0.05 |
ENST00000377275.3
|
ARL5B
|
ADP-ribosylation factor-like 5B |
| chr3_-_100120223 | 0.05 |
ENST00000284320.5
|
TOMM70A
|
translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) |
| chr3_+_152017181 | 0.05 |
ENST00000498502.1
ENST00000324196.5 ENST00000545754.1 ENST00000357472.3 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr5_+_65222299 | 0.05 |
ENST00000284037.5
|
ERBB2IP
|
erbb2 interacting protein |
| chr2_+_109335929 | 0.05 |
ENST00000283195.6
|
RANBP2
|
RAN binding protein 2 |
| chr9_+_4985228 | 0.05 |
ENST00000381652.3
|
JAK2
|
Janus kinase 2 |
| chr2_-_172017343 | 0.05 |
ENST00000431350.2
ENST00000360843.3 |
TLK1
|
tousled-like kinase 1 |
| chr5_-_127873659 | 0.05 |
ENST00000262464.4
|
FBN2
|
fibrillin 2 |
| chr3_+_88188254 | 0.05 |
ENST00000309495.5
|
ZNF654
|
zinc finger protein 654 |
| chr1_+_32479430 | 0.05 |
ENST00000327300.7
ENST00000492989.1 |
KHDRBS1
|
KH domain containing, RNA binding, signal transduction associated 1 |
| chr10_+_72575643 | 0.05 |
ENST00000373202.3
|
SGPL1
|
sphingosine-1-phosphate lyase 1 |
| chr6_+_71276596 | 0.04 |
ENST00000370474.3
|
C6orf57
|
chromosome 6 open reading frame 57 |
| chr2_-_73053126 | 0.04 |
ENST00000272427.6
ENST00000410104.1 |
EXOC6B
|
exocyst complex component 6B |
| chr15_-_68724490 | 0.04 |
ENST00000315757.7
ENST00000423218.2 |
ITGA11
|
integrin, alpha 11 |
| chr1_-_209979375 | 0.04 |
ENST00000367021.3
|
IRF6
|
interferon regulatory factor 6 |
| chr12_+_52203789 | 0.04 |
ENST00000599343.1
|
AC068987.1
|
HCG1997999; cDNA FLJ33996 fis, clone DFNES2008881 |
| chr5_-_131132614 | 0.04 |
ENST00000307968.7
ENST00000307954.8 |
FNIP1
|
folliculin interacting protein 1 |
| chr10_-_79686284 | 0.04 |
ENST00000372391.2
ENST00000372388.2 |
DLG5
|
discs, large homolog 5 (Drosophila) |
| chr7_-_105029329 | 0.04 |
ENST00000393651.3
ENST00000460391.1 |
SRPK2
|
SRSF protein kinase 2 |
| chr19_+_36486078 | 0.04 |
ENST00000378887.2
|
SDHAF1
|
succinate dehydrogenase complex assembly factor 1 |
| chr14_+_102196739 | 0.04 |
ENST00000556973.1
|
RP11-796G6.2
|
Uncharacterized protein |
| chr12_-_109125285 | 0.03 |
ENST00000552871.1
ENST00000261401.3 |
CORO1C
|
coronin, actin binding protein, 1C |
| chr19_-_474880 | 0.03 |
ENST00000382696.3
ENST00000315489.4 |
ODF3L2
|
outer dense fiber of sperm tails 3-like 2 |
| chr8_-_42751820 | 0.03 |
ENST00000526349.1
ENST00000527424.1 ENST00000534961.1 ENST00000319073.4 |
RNF170
|
ring finger protein 170 |
| chr6_-_86352642 | 0.03 |
ENST00000355238.6
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr12_+_110437328 | 0.03 |
ENST00000261739.4
|
ANKRD13A
|
ankyrin repeat domain 13A |
| chr1_+_90098606 | 0.03 |
ENST00000370454.4
|
LRRC8C
|
leucine rich repeat containing 8 family, member C |
| chr6_-_8064567 | 0.03 |
ENST00000543936.1
ENST00000397457.2 |
BLOC1S5
|
biogenesis of lysosomal organelles complex-1, subunit 5, muted |
| chr13_-_45915221 | 0.03 |
ENST00000309246.5
ENST00000379060.4 ENST00000379055.1 ENST00000527226.1 ENST00000379056.1 |
TPT1
|
tumor protein, translationally-controlled 1 |
| chr1_-_9970227 | 0.03 |
ENST00000377263.1
|
CTNNBIP1
|
catenin, beta interacting protein 1 |
| chr10_-_75910789 | 0.03 |
ENST00000355264.4
|
AP3M1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr11_-_6255846 | 0.03 |
ENST00000449352.2
|
FAM160A2
|
family with sequence similarity 160, member A2 |
| chr12_-_111021110 | 0.03 |
ENST00000354300.3
|
PPTC7
|
PTC7 protein phosphatase homolog (S. cerevisiae) |
| chr5_-_56247935 | 0.03 |
ENST00000381199.3
ENST00000381226.3 ENST00000381213.3 |
MIER3
|
mesoderm induction early response 1, family member 3 |
| chr3_-_171178157 | 0.03 |
ENST00000465393.1
ENST00000436636.2 ENST00000369326.5 ENST00000538048.1 ENST00000341852.6 |
TNIK
|
TRAF2 and NCK interacting kinase |
| chr2_+_148778570 | 0.03 |
ENST00000407073.1
|
MBD5
|
methyl-CpG binding domain protein 5 |
| chr10_+_88516396 | 0.03 |
ENST00000372037.3
|
BMPR1A
|
bone morphogenetic protein receptor, type IA |
| chr12_+_123868320 | 0.03 |
ENST00000402868.3
ENST00000330479.4 |
SETD8
|
SET domain containing (lysine methyltransferase) 8 |
| chr10_+_111967345 | 0.03 |
ENST00000332674.5
ENST00000453116.1 |
MXI1
|
MAX interactor 1, dimerization protein |
| chr9_-_140196703 | 0.02 |
ENST00000356628.2
|
NRARP
|
NOTCH-regulated ankyrin repeat protein |
| chr10_+_99344104 | 0.02 |
ENST00000555577.1
ENST00000370649.3 |
PI4K2A
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha Phosphatidylinositol 4-kinase type 2-alpha; Uncharacterized protein |
| chr5_+_172261228 | 0.02 |
ENST00000393784.3
|
ERGIC1
|
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
| chrX_-_119694538 | 0.02 |
ENST00000371322.5
|
CUL4B
|
cullin 4B |
| chr14_+_23775971 | 0.02 |
ENST00000250405.5
|
BCL2L2
|
BCL2-like 2 |
| chr10_-_14590644 | 0.02 |
ENST00000378470.1
|
FAM107B
|
family with sequence similarity 107, member B |
| chr11_+_111957497 | 0.02 |
ENST00000375549.3
ENST00000528182.1 ENST00000528048.1 ENST00000528021.1 ENST00000526592.1 ENST00000525291.1 |
SDHD
|
succinate dehydrogenase complex, subunit D, integral membrane protein |
| chr1_+_174769006 | 0.02 |
ENST00000489615.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chrX_-_25034065 | 0.02 |
ENST00000379044.4
|
ARX
|
aristaless related homeobox |
| chr3_+_132136331 | 0.02 |
ENST00000260818.6
|
DNAJC13
|
DnaJ (Hsp40) homolog, subfamily C, member 13 |
| chr7_-_158622210 | 0.02 |
ENST00000251527.5
|
ESYT2
|
extended synaptotagmin-like protein 2 |
| chr8_-_101965146 | 0.02 |
ENST00000395957.2
ENST00000395948.2 ENST00000457309.1 |
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr18_-_60987220 | 0.01 |
ENST00000398117.1
|
BCL2
|
B-cell CLL/lymphoma 2 |
| chr6_+_11094266 | 0.01 |
ENST00000416247.2
|
SMIM13
|
small integral membrane protein 13 |
| chr11_+_114270752 | 0.01 |
ENST00000540163.1
|
RBM7
|
RNA binding motif protein 7 |
| chr20_-_656823 | 0.01 |
ENST00000246104.6
|
SCRT2
|
scratch family zinc finger 2 |
| chr17_+_61699766 | 0.01 |
ENST00000579585.1
ENST00000584573.1 ENST00000361733.3 ENST00000361357.3 |
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3 |
| chr2_-_105946491 | 0.01 |
ENST00000393359.2
|
TGFBRAP1
|
transforming growth factor, beta receptor associated protein 1 |
| chr10_-_61666267 | 0.01 |
ENST00000263102.6
|
CCDC6
|
coiled-coil domain containing 6 |
| chr11_+_925824 | 0.01 |
ENST00000525796.1
ENST00000534328.1 ENST00000448903.2 ENST00000332231.5 |
AP2A2
|
adaptor-related protein complex 2, alpha 2 subunit |
| chr16_+_87425381 | 0.01 |
ENST00000268607.5
|
MAP1LC3B
|
microtubule-associated protein 1 light chain 3 beta |
| chr9_+_133454943 | 0.01 |
ENST00000319725.9
|
FUBP3
|
far upstream element (FUSE) binding protein 3 |
| chr15_+_41523335 | 0.01 |
ENST00000334660.5
|
CHP1
|
calcineurin-like EF-hand protein 1 |
| chr16_-_71758602 | 0.01 |
ENST00000568954.1
|
PHLPP2
|
PH domain and leucine rich repeat protein phosphatase 2 |
| chr7_-_140098257 | 0.01 |
ENST00000340308.3
ENST00000447932.2 ENST00000429996.2 ENST00000469193.1 ENST00000326232.9 |
SLC37A3
|
solute carrier family 37, member 3 |
| chr14_-_27066636 | 0.01 |
ENST00000267422.7
ENST00000344429.5 ENST00000574031.1 ENST00000465357.2 ENST00000547619.1 |
NOVA1
|
neuro-oncological ventral antigen 1 |
| chr2_-_176032843 | 0.01 |
ENST00000392544.1
ENST00000409499.1 ENST00000426833.3 ENST00000392543.2 ENST00000538946.1 ENST00000487334.2 ENST00000409833.1 ENST00000409635.1 ENST00000264110.2 ENST00000345739.5 |
ATF2
|
activating transcription factor 2 |
| chr1_+_36348790 | 0.01 |
ENST00000373204.4
|
AGO1
|
argonaute RISC catalytic component 1 |
| chr1_+_16693578 | 0.01 |
ENST00000401088.4
ENST00000471507.1 ENST00000401089.3 ENST00000375590.3 ENST00000492354.1 |
SZRD1
|
SUZ RNA binding domain containing 1 |
| chr6_+_114178512 | 0.01 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr3_+_196295482 | 0.00 |
ENST00000440469.1
ENST00000311630.6 |
FBXO45
|
F-box protein 45 |
| chr7_+_138916231 | 0.00 |
ENST00000473989.3
ENST00000288561.8 |
UBN2
|
ubinuclein 2 |
| chr1_+_32930647 | 0.00 |
ENST00000609129.1
|
ZBTB8B
|
zinc finger and BTB domain containing 8B |
| chr16_+_70148230 | 0.00 |
ENST00000398122.3
ENST00000568530.1 |
PDPR
|
pyruvate dehydrogenase phosphatase regulatory subunit |
Network of associatons between targets according to the STRING database.
First level regulatory network of UCCCUUU
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1990619 | histone H3-K9 deacetylation(GO:1990619) |
| 0.1 | 0.3 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.1 | 0.3 | GO:0003274 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) endocardial cushion fusion(GO:0003274) |
| 0.1 | 0.3 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.1 | 0.2 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.1 | 0.3 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 0.2 | GO:1904640 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) response to methionine(GO:1904640) |
| 0.1 | 0.4 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.3 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.3 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.1 | GO:0046963 | 3'-phosphoadenosine 5'-phosphosulfate transport(GO:0046963) 3'-phospho-5'-adenylyl sulfate transmembrane transport(GO:1902559) |
| 0.0 | 0.2 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.0 | 0.4 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.1 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.0 | 0.1 | GO:0060995 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.0 | 0.2 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.0 | 0.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.4 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:0042495 | detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.0 | 0.1 | GO:1902513 | regulation of organelle transport along microtubule(GO:1902513) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.0 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.1 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.2 | GO:1901725 | regulation of histone deacetylase activity(GO:1901725) |
| 0.0 | 0.0 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.1 | 0.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.2 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.2 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.4 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.1 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.1 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.0 | GO:0034681 | integrin alpha11-beta1 complex(GO:0034681) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.1 | 0.3 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.1 | 0.3 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.1 | 0.3 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.1 | 0.4 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.1 | 0.3 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 0.2 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.4 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.1 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.0 | 0.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.1 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.1 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | ST STAT3 PATHWAY | STAT3 Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |