Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
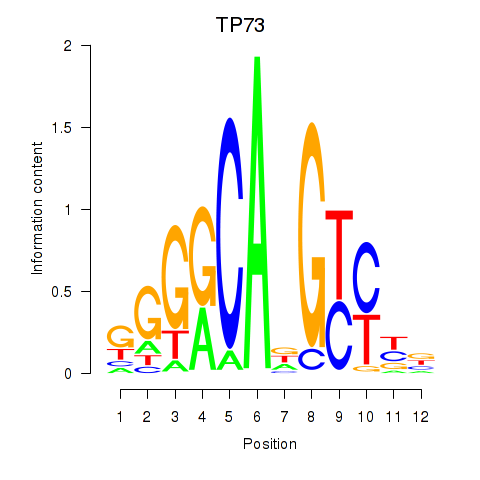
Results for TP73
Z-value: 0.74
Transcription factors associated with TP73
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TP73
|
ENSG00000078900.10 | tumor protein p73 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TP73 | hg19_v2_chr1_+_3569129_3569150 | 0.65 | 3.5e-01 | Click! |
Activity profile of TP73 motif
Sorted Z-values of TP73 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_62203808 | 1.31 |
ENST00000467148.1
|
HELZ2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr20_+_35504522 | 0.89 |
ENST00000602922.1
ENST00000217320.3 |
TLDC2
|
TBC/LysM-associated domain containing 2 |
| chr17_+_73750699 | 0.69 |
ENST00000584939.1
|
ITGB4
|
integrin, beta 4 |
| chr3_+_136676707 | 0.57 |
ENST00000329582.4
|
IL20RB
|
interleukin 20 receptor beta |
| chr20_+_48807351 | 0.53 |
ENST00000303004.3
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta |
| chr11_+_65154070 | 0.52 |
ENST00000317568.5
ENST00000531296.1 ENST00000533782.1 ENST00000355991.5 ENST00000416776.2 ENST00000526201.1 |
FRMD8
|
FERM domain containing 8 |
| chr15_+_74466744 | 0.49 |
ENST00000560862.1
ENST00000395118.1 |
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr3_+_136676851 | 0.46 |
ENST00000309741.5
|
IL20RB
|
interleukin 20 receptor beta |
| chr16_+_8891670 | 0.43 |
ENST00000268261.4
ENST00000539622.1 ENST00000569958.1 ENST00000537352.1 |
PMM2
|
phosphomannomutase 2 |
| chr10_-_99185798 | 0.38 |
ENST00000439965.2
|
AL355490.1
|
LOC644215 protein; Uncharacterized protein |
| chr10_-_47173994 | 0.36 |
ENST00000414655.2
ENST00000545298.1 ENST00000359178.4 ENST00000358140.4 ENST00000503031.1 |
ANXA8L1
LINC00842
|
annexin A8-like 1 long intergenic non-protein coding RNA 842 |
| chr22_-_20255212 | 0.35 |
ENST00000416372.1
|
RTN4R
|
reticulon 4 receptor |
| chr15_+_57891609 | 0.32 |
ENST00000569089.1
|
MYZAP
|
myocardial zonula adherens protein |
| chr17_-_42452063 | 0.32 |
ENST00000588098.1
|
ITGA2B
|
integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) |
| chr10_+_47746929 | 0.32 |
ENST00000340243.6
ENST00000374277.5 ENST00000449464.2 ENST00000538825.1 ENST00000335083.5 |
ANXA8L2
AL603965.1
|
annexin A8-like 2 Protein LOC100996760 |
| chr22_+_39966758 | 0.31 |
ENST00000407673.1
ENST00000401624.1 ENST00000404898.1 ENST00000402142.3 ENST00000336649.4 ENST00000400164.3 |
CACNA1I
|
calcium channel, voltage-dependent, T type, alpha 1I subunit |
| chr2_-_26864228 | 0.29 |
ENST00000288861.4
|
CIB4
|
calcium and integrin binding family member 4 |
| chr7_-_100493482 | 0.29 |
ENST00000411582.1
ENST00000419336.2 ENST00000241069.5 ENST00000302913.4 |
ACHE
|
acetylcholinesterase (Yt blood group) |
| chr2_-_230135937 | 0.29 |
ENST00000392054.3
ENST00000409462.1 ENST00000392055.3 |
PID1
|
phosphotyrosine interaction domain containing 1 |
| chr8_+_145065521 | 0.23 |
ENST00000534791.1
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr6_-_33256664 | 0.22 |
ENST00000444176.1
|
WDR46
|
WD repeat domain 46 |
| chr5_+_125758813 | 0.22 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr17_-_40835076 | 0.22 |
ENST00000591765.1
|
CCR10
|
chemokine (C-C motif) receptor 10 |
| chr5_+_125758865 | 0.21 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr15_-_74043816 | 0.21 |
ENST00000379822.4
|
C15orf59
|
chromosome 15 open reading frame 59 |
| chr15_+_75498739 | 0.21 |
ENST00000565074.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr10_-_105238997 | 0.20 |
ENST00000369783.4
|
CALHM3
|
calcium homeostasis modulator 3 |
| chrX_-_80377162 | 0.20 |
ENST00000430960.1
ENST00000447319.1 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr7_-_100171270 | 0.20 |
ENST00000538735.1
|
SAP25
|
Sin3A-associated protein, 25kDa |
| chr12_-_82752159 | 0.20 |
ENST00000552377.1
|
CCDC59
|
coiled-coil domain containing 59 |
| chr17_+_43318434 | 0.20 |
ENST00000587489.1
|
FMNL1
|
formin-like 1 |
| chr1_-_78444776 | 0.19 |
ENST00000370767.1
ENST00000421641.1 |
FUBP1
|
far upstream element (FUSE) binding protein 1 |
| chr8_+_145065705 | 0.18 |
ENST00000533044.1
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr1_-_78444738 | 0.18 |
ENST00000436586.2
ENST00000370768.2 |
FUBP1
|
far upstream element (FUSE) binding protein 1 |
| chr9_+_6758109 | 0.18 |
ENST00000536108.1
|
KDM4C
|
lysine (K)-specific demethylase 4C |
| chr3_+_48481658 | 0.18 |
ENST00000438607.2
|
TMA7
|
translation machinery associated 7 homolog (S. cerevisiae) |
| chr10_-_97200772 | 0.18 |
ENST00000371241.1
ENST00000354106.3 ENST00000371239.1 ENST00000361941.3 ENST00000277982.5 ENST00000371245.3 |
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr14_-_75078725 | 0.18 |
ENST00000556690.1
|
LTBP2
|
latent transforming growth factor beta binding protein 2 |
| chr15_+_75498355 | 0.18 |
ENST00000567617.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr15_+_41136369 | 0.17 |
ENST00000563656.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr15_+_41136263 | 0.17 |
ENST00000568823.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr14_+_103566665 | 0.17 |
ENST00000559116.1
|
EXOC3L4
|
exocyst complex component 3-like 4 |
| chr9_-_128003606 | 0.17 |
ENST00000324460.6
|
HSPA5
|
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) |
| chr14_+_105935396 | 0.16 |
ENST00000494981.1
|
MTA1
|
metastasis associated 1 |
| chr19_+_45254529 | 0.15 |
ENST00000444487.1
|
BCL3
|
B-cell CLL/lymphoma 3 |
| chr8_+_54764346 | 0.15 |
ENST00000297313.3
ENST00000344277.6 |
RGS20
|
regulator of G-protein signaling 20 |
| chr2_+_220283091 | 0.15 |
ENST00000373960.3
|
DES
|
desmin |
| chr16_+_67197288 | 0.15 |
ENST00000264009.8
ENST00000421453.1 |
HSF4
|
heat shock transcription factor 4 |
| chr5_+_43120985 | 0.15 |
ENST00000515326.1
|
ZNF131
|
zinc finger protein 131 |
| chr17_+_37356528 | 0.14 |
ENST00000225430.4
|
RPL19
|
ribosomal protein L19 |
| chr9_+_6758024 | 0.14 |
ENST00000442236.2
|
KDM4C
|
lysine (K)-specific demethylase 4C |
| chr9_+_6757634 | 0.13 |
ENST00000543771.1
ENST00000401787.3 ENST00000381306.3 ENST00000381309.3 |
KDM4C
|
lysine (K)-specific demethylase 4C |
| chr16_+_84178874 | 0.13 |
ENST00000378553.5
|
DNAAF1
|
dynein, axonemal, assembly factor 1 |
| chr1_-_68698222 | 0.13 |
ENST00000370976.3
ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr15_+_64680003 | 0.13 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr16_-_84178728 | 0.13 |
ENST00000562224.1
ENST00000434463.3 ENST00000564998.1 ENST00000219439.4 |
HSDL1
|
hydroxysteroid dehydrogenase like 1 |
| chr3_+_100120441 | 0.12 |
ENST00000489752.1
|
LNP1
|
leukemia NUP98 fusion partner 1 |
| chr17_+_72427477 | 0.11 |
ENST00000342648.5
ENST00000481232.1 |
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr2_-_232328867 | 0.11 |
ENST00000453992.1
ENST00000417652.1 ENST00000454824.1 |
NCL
|
nucleolin |
| chr19_+_49622646 | 0.11 |
ENST00000334186.4
|
PPFIA3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr14_-_25519317 | 0.10 |
ENST00000323944.5
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr20_-_52790512 | 0.10 |
ENST00000216862.3
|
CYP24A1
|
cytochrome P450, family 24, subfamily A, polypeptide 1 |
| chr6_+_155538093 | 0.10 |
ENST00000462408.2
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr7_-_2883928 | 0.10 |
ENST00000275364.3
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12 |
| chr8_-_70745575 | 0.09 |
ENST00000524945.1
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1 |
| chr6_-_112194484 | 0.09 |
ENST00000518295.1
ENST00000484067.2 ENST00000229470.5 ENST00000356013.2 ENST00000368678.4 ENST00000523238.1 ENST00000354650.3 |
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr8_-_145016692 | 0.09 |
ENST00000357649.2
|
PLEC
|
plectin |
| chr7_-_27213893 | 0.09 |
ENST00000283921.4
|
HOXA10
|
homeobox A10 |
| chr5_+_125759140 | 0.09 |
ENST00000543198.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr16_+_75032901 | 0.09 |
ENST00000335325.4
ENST00000320619.6 |
ZNRF1
|
zinc and ring finger 1, E3 ubiquitin protein ligase |
| chr18_+_77623668 | 0.09 |
ENST00000316249.3
|
KCNG2
|
potassium voltage-gated channel, subfamily G, member 2 |
| chr14_-_25519095 | 0.09 |
ENST00000419632.2
ENST00000358326.2 ENST00000396700.1 ENST00000548724.1 |
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr20_-_49253425 | 0.09 |
ENST00000045083.2
|
FAM65C
|
family with sequence similarity 65, member C |
| chr4_+_6717842 | 0.08 |
ENST00000320776.3
|
BLOC1S4
|
biogenesis of lysosomal organelles complex-1, subunit 4, cappuccino |
| chr2_-_158485387 | 0.08 |
ENST00000243349.8
|
ACVR1C
|
activin A receptor, type IC |
| chr16_+_84178850 | 0.08 |
ENST00000334315.5
|
DNAAF1
|
dynein, axonemal, assembly factor 1 |
| chr1_+_47137544 | 0.08 |
ENST00000564373.1
|
TEX38
|
testis expressed 38 |
| chr1_+_156785425 | 0.07 |
ENST00000392302.2
|
NTRK1
|
neurotrophic tyrosine kinase, receptor, type 1 |
| chr17_-_47492236 | 0.07 |
ENST00000434917.2
ENST00000300408.3 ENST00000511832.1 ENST00000419140.2 |
PHB
|
prohibitin |
| chr2_-_73511559 | 0.07 |
ENST00000521871.1
|
FBXO41
|
F-box protein 41 |
| chr20_-_52790055 | 0.07 |
ENST00000395955.3
|
CYP24A1
|
cytochrome P450, family 24, subfamily A, polypeptide 1 |
| chr12_-_82752565 | 0.07 |
ENST00000256151.7
|
CCDC59
|
coiled-coil domain containing 59 |
| chr1_+_162467595 | 0.07 |
ENST00000538489.1
ENST00000489294.1 |
UHMK1
|
U2AF homology motif (UHM) kinase 1 |
| chr17_-_79818354 | 0.07 |
ENST00000576541.1
ENST00000576380.1 ENST00000571617.1 ENST00000576052.1 ENST00000576390.1 ENST00000573778.2 ENST00000439918.2 ENST00000574914.1 ENST00000331483.4 |
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr11_-_17035943 | 0.06 |
ENST00000355661.3
ENST00000532079.1 ENST00000448080.2 ENST00000531066.1 |
PLEKHA7
|
pleckstrin homology domain containing, family A member 7 |
| chr1_-_52344471 | 0.06 |
ENST00000352171.7
ENST00000354831.7 |
NRD1
|
nardilysin (N-arginine dibasic convertase) |
| chr19_-_35981358 | 0.06 |
ENST00000484218.2
ENST00000338897.3 |
KRTDAP
|
keratinocyte differentiation-associated protein |
| chr15_+_41136216 | 0.06 |
ENST00000562057.1
ENST00000344051.4 |
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr11_-_31833719 | 0.06 |
ENST00000524853.1
|
PAX6
|
paired box 6 |
| chr18_-_4455260 | 0.05 |
ENST00000581527.1
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr12_+_96196875 | 0.05 |
ENST00000553095.1
|
RP11-536G4.1
|
Uncharacterized protein |
| chr2_-_27357479 | 0.05 |
ENST00000406567.3
ENST00000260643.2 |
PREB
|
prolactin regulatory element binding |
| chr16_+_222846 | 0.05 |
ENST00000251595.6
ENST00000397806.1 |
HBA2
|
hemoglobin, alpha 2 |
| chr19_-_39226045 | 0.05 |
ENST00000597987.1
ENST00000595177.1 |
CAPN12
|
calpain 12 |
| chr1_+_47137445 | 0.05 |
ENST00000569393.1
ENST00000334122.4 ENST00000415500.1 |
TEX38
|
testis expressed 38 |
| chr17_+_37356586 | 0.05 |
ENST00000579260.1
ENST00000582193.1 |
RPL19
|
ribosomal protein L19 |
| chr17_-_2615031 | 0.05 |
ENST00000576885.1
ENST00000574426.2 |
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr7_-_128045984 | 0.04 |
ENST00000470772.1
ENST00000480861.1 ENST00000496200.1 |
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1 |
| chr11_+_236959 | 0.04 |
ENST00000431206.2
ENST00000528906.1 |
PSMD13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr17_+_37356555 | 0.04 |
ENST00000579374.1
|
RPL19
|
ribosomal protein L19 |
| chr19_+_19627026 | 0.04 |
ENST00000608404.1
ENST00000555938.1 ENST00000503283.1 ENST00000512771.3 ENST00000428459.2 |
YJEFN3
CTC-260F20.3
NDUFA13
|
YjeF N-terminal domain containing 3 Uncharacterized protein NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13 |
| chr17_-_2614927 | 0.04 |
ENST00000435359.1
|
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chrX_-_138724994 | 0.04 |
ENST00000536274.1
|
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr9_+_87285257 | 0.04 |
ENST00000323115.4
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr7_-_100493744 | 0.04 |
ENST00000428317.1
ENST00000441605.1 |
ACHE
|
acetylcholinesterase (Yt blood group) |
| chr9_-_130517309 | 0.04 |
ENST00000414380.1
|
SH2D3C
|
SH2 domain containing 3C |
| chr16_+_67195592 | 0.04 |
ENST00000519378.1
|
FBXL8
|
F-box and leucine-rich repeat protein 8 |
| chr17_-_47492164 | 0.04 |
ENST00000512041.2
ENST00000446735.1 ENST00000504124.1 |
PHB
|
prohibitin |
| chrX_-_80377118 | 0.03 |
ENST00000373250.3
|
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chrX_-_138724677 | 0.03 |
ENST00000370573.4
ENST00000338585.6 ENST00000370576.4 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr1_-_236228403 | 0.03 |
ENST00000366595.3
|
NID1
|
nidogen 1 |
| chr1_-_68698197 | 0.03 |
ENST00000370973.2
ENST00000370971.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr9_+_133223932 | 0.03 |
ENST00000420499.2
|
HMCN2
|
hemicentin 2 |
| chr2_+_95940220 | 0.03 |
ENST00000542147.1
|
PROM2
|
prominin 2 |
| chr16_-_14724057 | 0.03 |
ENST00000539279.1
ENST00000420015.2 ENST00000437198.2 |
PARN
|
poly(A)-specific ribonuclease |
| chr15_+_41136586 | 0.03 |
ENST00000431806.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr16_-_1146244 | 0.03 |
ENST00000328449.5
|
C1QTNF8
|
C1q and tumor necrosis factor related protein 8 |
| chr17_+_78388959 | 0.02 |
ENST00000518137.1
ENST00000520367.1 ENST00000523999.1 ENST00000323854.5 ENST00000522751.1 |
ENDOV
|
endonuclease V |
| chr2_-_26205550 | 0.02 |
ENST00000405914.1
|
KIF3C
|
kinesin family member 3C |
| chr18_-_53253000 | 0.02 |
ENST00000566514.1
|
TCF4
|
transcription factor 4 |
| chr9_+_36169380 | 0.02 |
ENST00000335119.2
|
CCIN
|
calicin |
| chr1_+_110082487 | 0.02 |
ENST00000527748.1
|
GPR61
|
G protein-coupled receptor 61 |
| chr9_-_132383055 | 0.02 |
ENST00000372478.4
|
C9orf50
|
chromosome 9 open reading frame 50 |
| chr1_-_236228417 | 0.01 |
ENST00000264187.6
|
NID1
|
nidogen 1 |
| chr6_-_46703069 | 0.01 |
ENST00000538237.1
ENST00000274793.7 |
PLA2G7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr6_+_31515337 | 0.01 |
ENST00000376148.4
ENST00000376145.4 |
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chrX_+_9880590 | 0.01 |
ENST00000452575.1
|
SHROOM2
|
shroom family member 2 |
| chr12_+_113495492 | 0.01 |
ENST00000257600.3
|
DTX1
|
deltex homolog 1 (Drosophila) |
| chr11_+_66234216 | 0.01 |
ENST00000349459.6
ENST00000320740.7 ENST00000524466.1 ENST00000526296.1 |
PELI3
|
pellino E3 ubiquitin protein ligase family member 3 |
| chr19_-_19626838 | 0.01 |
ENST00000360913.3
|
TSSK6
|
testis-specific serine kinase 6 |
| chr18_-_53253323 | 0.01 |
ENST00000540999.1
ENST00000563888.2 |
TCF4
|
transcription factor 4 |
| chr10_-_105212059 | 0.01 |
ENST00000260743.5
|
CALHM2
|
calcium homeostasis modulator 2 |
| chr11_-_65686496 | 0.00 |
ENST00000449692.3
|
C11orf68
|
chromosome 11 open reading frame 68 |
| chr1_-_214724566 | 0.00 |
ENST00000366956.5
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14 |
| chr9_+_138391805 | 0.00 |
ENST00000371785.1
|
MRPS2
|
mitochondrial ribosomal protein S2 |
| chr17_-_39023462 | 0.00 |
ENST00000251643.4
|
KRT12
|
keratin 12 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TP73
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.1 | 0.3 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.1 | 0.4 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.3 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.1 | 0.2 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.0 | 0.7 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.2 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.0 | 0.5 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.7 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.2 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.2 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.0 | 0.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:0097360 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.0 | 0.4 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.1 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.4 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 0.4 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.4 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.1 | 0.3 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.3 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.1 | 0.3 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 0.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.7 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.2 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.4 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.5 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 1.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.0 | GO:0052857 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.2 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |