Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
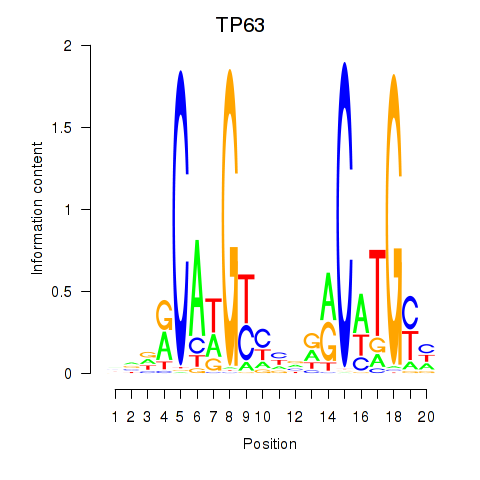
Results for TP63
Z-value: 0.45
Transcription factors associated with TP63
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TP63
|
ENSG00000073282.8 | tumor protein p63 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TP63 | hg19_v2_chr3_+_189507523_189507590 | 0.77 | 2.3e-01 | Click! |
Activity profile of TP63 motif
Sorted Z-values of TP63 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_90751038 | 0.39 |
ENST00000458159.1
ENST00000415557.1 ENST00000458208.1 |
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr10_-_88729200 | 0.35 |
ENST00000474994.2
|
MMRN2
|
multimerin 2 |
| chr12_+_69202795 | 0.32 |
ENST00000539479.1
ENST00000393415.3 ENST00000523991.1 ENST00000543323.1 ENST00000393416.2 |
MDM2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr1_-_38157877 | 0.30 |
ENST00000477060.1
ENST00000491981.1 ENST00000488137.1 |
C1orf109
|
chromosome 1 open reading frame 109 |
| chr16_+_57653854 | 0.25 |
ENST00000568908.1
ENST00000568909.1 ENST00000566778.1 ENST00000561988.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr5_+_162864575 | 0.23 |
ENST00000512163.1
ENST00000393929.1 ENST00000340828.2 ENST00000511683.2 ENST00000510097.1 ENST00000511490.2 ENST00000510664.1 |
CCNG1
|
cyclin G1 |
| chr10_-_88729069 | 0.22 |
ENST00000609457.1
|
MMRN2
|
multimerin 2 |
| chr16_+_57653989 | 0.20 |
ENST00000567835.1
ENST00000569372.1 ENST00000563548.1 ENST00000562003.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr2_+_113299990 | 0.19 |
ENST00000537335.1
ENST00000417433.2 |
POLR1B
|
polymerase (RNA) I polypeptide B, 128kDa |
| chr12_+_69202975 | 0.19 |
ENST00000544561.1
ENST00000393410.1 ENST00000299252.4 ENST00000360430.2 ENST00000517852.1 ENST00000545204.1 ENST00000393413.3 ENST00000350057.5 ENST00000348801.2 ENST00000478070.1 |
MDM2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr15_+_69857515 | 0.19 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr6_-_109330702 | 0.18 |
ENST00000356644.7
|
SESN1
|
sestrin 1 |
| chr9_+_124088860 | 0.18 |
ENST00000373806.1
|
GSN
|
gelsolin |
| chr22_+_39493207 | 0.17 |
ENST00000348946.4
ENST00000442487.3 ENST00000421988.2 |
APOBEC3H
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3H |
| chr3_+_44840679 | 0.17 |
ENST00000425755.1
|
KIF15
|
kinesin family member 15 |
| chr5_+_115420688 | 0.17 |
ENST00000274458.4
|
COMMD10
|
COMM domain containing 10 |
| chr6_+_35265586 | 0.17 |
ENST00000542066.1
ENST00000316637.5 |
DEF6
|
differentially expressed in FDCP 6 homolog (mouse) |
| chr11_+_64073699 | 0.17 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr10_+_99344071 | 0.16 |
ENST00000370647.4
ENST00000370646.4 |
HOGA1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr5_+_27472371 | 0.16 |
ENST00000512067.1
ENST00000510165.1 ENST00000505775.1 ENST00000514844.1 ENST00000503107.1 |
LINC01021
|
long intergenic non-protein coding RNA 1021 |
| chr17_-_4938712 | 0.16 |
ENST00000254853.5
ENST00000424747.1 |
SLC52A1
|
solute carrier family 52 (riboflavin transporter), member 1 |
| chr7_+_100466433 | 0.15 |
ENST00000429658.1
|
TRIP6
|
thyroid hormone receptor interactor 6 |
| chr1_+_38158090 | 0.15 |
ENST00000373055.1
ENST00000327331.2 |
CDCA8
|
cell division cycle associated 8 |
| chr22_+_37959647 | 0.15 |
ENST00000415670.1
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chr3_-_123168551 | 0.15 |
ENST00000462833.1
|
ADCY5
|
adenylate cyclase 5 |
| chr7_+_79763271 | 0.15 |
ENST00000442586.1
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr1_-_220263096 | 0.15 |
ENST00000463953.1
ENST00000354807.3 ENST00000414869.2 ENST00000498237.2 ENST00000498791.2 ENST00000544404.1 ENST00000480959.2 ENST00000322067.7 |
BPNT1
|
3'(2'), 5'-bisphosphate nucleotidase 1 |
| chr7_+_140396946 | 0.15 |
ENST00000476470.1
ENST00000471136.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr16_-_47493041 | 0.15 |
ENST00000565940.2
|
ITFG1
|
integrin alpha FG-GAP repeat containing 1 |
| chr19_+_42364460 | 0.14 |
ENST00000593863.1
|
RPS19
|
ribosomal protein S19 |
| chr11_+_64950801 | 0.14 |
ENST00000526468.1
|
CAPN1
|
calpain 1, (mu/I) large subunit |
| chr19_-_47734448 | 0.14 |
ENST00000439096.2
|
BBC3
|
BCL2 binding component 3 |
| chr3_-_167813672 | 0.14 |
ENST00000470487.1
|
GOLIM4
|
golgi integral membrane protein 4 |
| chr17_+_4618734 | 0.14 |
ENST00000571206.1
|
ARRB2
|
arrestin, beta 2 |
| chr1_+_42619070 | 0.14 |
ENST00000372581.1
|
GUCA2B
|
guanylate cyclase activator 2B (uroguanylin) |
| chr2_-_216003127 | 0.14 |
ENST00000412081.1
ENST00000272895.7 |
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr15_-_63448973 | 0.14 |
ENST00000462430.1
|
RPS27L
|
ribosomal protein S27-like |
| chr1_-_19229014 | 0.13 |
ENST00000538839.1
ENST00000290597.5 |
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr10_+_99344104 | 0.13 |
ENST00000555577.1
ENST00000370649.3 |
PI4K2A
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha Phosphatidylinositol 4-kinase type 2-alpha; Uncharacterized protein |
| chr9_+_35906176 | 0.13 |
ENST00000354323.2
|
HRCT1
|
histidine rich carboxyl terminus 1 |
| chr3_+_23986748 | 0.13 |
ENST00000312521.4
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr11_-_75921780 | 0.12 |
ENST00000529461.1
|
WNT11
|
wingless-type MMTV integration site family, member 11 |
| chr15_-_90233866 | 0.12 |
ENST00000561257.1
|
PEX11A
|
peroxisomal biogenesis factor 11 alpha |
| chr4_+_3388057 | 0.12 |
ENST00000538395.1
|
RGS12
|
regulator of G-protein signaling 12 |
| chr22_+_29168652 | 0.12 |
ENST00000249064.4
ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117
|
coiled-coil domain containing 117 |
| chr7_+_140396756 | 0.12 |
ENST00000460088.1
ENST00000472695.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr16_+_10479906 | 0.12 |
ENST00000562527.1
ENST00000396560.2 ENST00000396559.1 ENST00000562102.1 ENST00000543967.1 ENST00000569939.1 ENST00000569900.1 |
ATF7IP2
|
activating transcription factor 7 interacting protein 2 |
| chr6_-_44923160 | 0.11 |
ENST00000371458.1
|
SUPT3H
|
suppressor of Ty 3 homolog (S. cerevisiae) |
| chr19_-_19887185 | 0.11 |
ENST00000590092.1
ENST00000589910.1 ENST00000589508.1 ENST00000586645.1 ENST00000586816.1 ENST00000588223.1 ENST00000586885.1 |
LINC00663
|
long intergenic non-protein coding RNA 663 |
| chr15_+_32907691 | 0.11 |
ENST00000361627.3
ENST00000567348.1 ENST00000563864.1 ENST00000543522.1 |
ARHGAP11A
|
Rho GTPase activating protein 11A |
| chr1_+_24645915 | 0.11 |
ENST00000350501.5
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr1_+_24645865 | 0.11 |
ENST00000342072.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr7_-_55620433 | 0.11 |
ENST00000418904.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr8_+_117778736 | 0.11 |
ENST00000309822.2
ENST00000357148.3 ENST00000517814.1 ENST00000517820.1 |
UTP23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chr9_+_91606355 | 0.11 |
ENST00000358157.2
|
S1PR3
|
sphingosine-1-phosphate receptor 3 |
| chr1_+_24645807 | 0.11 |
ENST00000361548.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr1_-_19229248 | 0.11 |
ENST00000375341.3
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr4_-_144826682 | 0.11 |
ENST00000358615.4
ENST00000437468.2 |
GYPE
|
glycophorin E (MNS blood group) |
| chr14_+_24779340 | 0.11 |
ENST00000533293.1
ENST00000543919.1 |
LTB4R2
|
leukotriene B4 receptor 2 |
| chr7_-_110174754 | 0.10 |
ENST00000435466.1
|
AC003088.1
|
AC003088.1 |
| chr1_+_24646002 | 0.10 |
ENST00000356046.2
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr17_+_26662679 | 0.10 |
ENST00000578158.1
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr3_-_57113314 | 0.10 |
ENST00000338458.4
ENST00000468727.1 |
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr6_-_101329191 | 0.10 |
ENST00000324723.6
ENST00000369162.2 ENST00000522650.1 |
ASCC3
|
activating signal cointegrator 1 complex subunit 3 |
| chr13_+_48611665 | 0.10 |
ENST00000258662.2
|
NUDT15
|
nudix (nucleoside diphosphate linked moiety X)-type motif 15 |
| chr17_-_70989072 | 0.10 |
ENST00000582769.1
|
SLC39A11
|
solute carrier family 39, member 11 |
| chr11_-_77899634 | 0.10 |
ENST00000526208.1
ENST00000529350.1 ENST00000530018.1 ENST00000528776.1 |
KCTD21
|
potassium channel tetramerization domain containing 21 |
| chrX_+_55478538 | 0.10 |
ENST00000342972.1
|
MAGEH1
|
melanoma antigen family H, 1 |
| chr15_+_68570062 | 0.09 |
ENST00000306917.4
|
FEM1B
|
fem-1 homolog b (C. elegans) |
| chr7_-_99869799 | 0.09 |
ENST00000436886.2
|
GATS
|
GATS, stromal antigen 3 opposite strand |
| chr15_+_69365272 | 0.09 |
ENST00000559914.1
ENST00000558369.1 |
LINC00277
|
long intergenic non-protein coding RNA 277 |
| chr5_+_41904431 | 0.09 |
ENST00000381647.2
|
C5orf51
|
chromosome 5 open reading frame 51 |
| chr19_+_42364313 | 0.09 |
ENST00000601492.1
ENST00000600467.1 ENST00000221975.2 |
RPS19
|
ribosomal protein S19 |
| chr12_-_88974236 | 0.09 |
ENST00000228280.5
ENST00000552044.1 ENST00000357116.4 |
KITLG
|
KIT ligand |
| chr1_+_114447763 | 0.09 |
ENST00000369563.3
|
DCLRE1B
|
DNA cross-link repair 1B |
| chr11_-_111782696 | 0.09 |
ENST00000227251.3
ENST00000526180.1 |
CRYAB
|
crystallin, alpha B |
| chr19_+_49458107 | 0.09 |
ENST00000539787.1
ENST00000345358.7 ENST00000391871.3 ENST00000415969.2 ENST00000354470.3 ENST00000506183.1 ENST00000293288.8 |
BAX
|
BCL2-associated X protein |
| chr19_-_47288162 | 0.09 |
ENST00000594991.1
|
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr22_-_38577782 | 0.09 |
ENST00000430886.1
ENST00000332509.3 ENST00000447598.2 ENST00000435484.1 ENST00000402064.1 ENST00000436218.1 |
PLA2G6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr4_-_13629269 | 0.09 |
ENST00000040738.5
|
BOD1L1
|
biorientation of chromosomes in cell division 1-like 1 |
| chr4_+_671711 | 0.09 |
ENST00000400159.2
|
MYL5
|
myosin, light chain 5, regulatory |
| chrX_+_153029633 | 0.09 |
ENST00000538966.1
ENST00000361971.5 ENST00000538776.1 ENST00000538543.1 |
PLXNB3
|
plexin B3 |
| chr19_+_19779589 | 0.09 |
ENST00000541458.1
|
ZNF101
|
zinc finger protein 101 |
| chr11_-_64889649 | 0.09 |
ENST00000434372.2
|
FAU
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed |
| chr11_-_64889252 | 0.09 |
ENST00000525297.1
ENST00000529259.1 |
FAU
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed |
| chr11_-_64889529 | 0.08 |
ENST00000531743.1
ENST00000527548.1 ENST00000526555.1 ENST00000279259.3 |
FAU
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed |
| chr3_+_184081137 | 0.08 |
ENST00000443489.1
|
POLR2H
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr11_-_71823266 | 0.08 |
ENST00000538919.1
ENST00000539395.1 ENST00000542531.1 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr3_+_45017722 | 0.08 |
ENST00000265564.7
|
EXOSC7
|
exosome component 7 |
| chr2_+_217524323 | 0.08 |
ENST00000456764.1
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa |
| chr1_-_93426998 | 0.08 |
ENST00000370310.4
|
FAM69A
|
family with sequence similarity 69, member A |
| chr1_+_85527987 | 0.08 |
ENST00000326813.8
ENST00000294664.6 ENST00000528899.1 |
WDR63
|
WD repeat domain 63 |
| chr16_+_19467772 | 0.08 |
ENST00000219821.5
ENST00000561503.1 ENST00000564959.1 |
TMC5
|
transmembrane channel-like 5 |
| chr15_-_63449663 | 0.08 |
ENST00000439025.1
|
RPS27L
|
ribosomal protein S27-like |
| chr5_-_42811986 | 0.08 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr19_+_19779686 | 0.08 |
ENST00000415784.2
|
ZNF101
|
zinc finger protein 101 |
| chr15_-_55700216 | 0.08 |
ENST00000569205.1
|
CCPG1
|
cell cycle progression 1 |
| chr1_+_15764931 | 0.08 |
ENST00000375949.4
ENST00000375943.2 |
CTRC
|
chymotrypsin C (caldecrin) |
| chr20_+_57264187 | 0.08 |
ENST00000525967.1
ENST00000525817.1 |
NPEPL1
|
aminopeptidase-like 1 |
| chr4_+_8321882 | 0.08 |
ENST00000509453.1
ENST00000503186.1 |
RP11-774O3.2
RP11-774O3.1
|
RP11-774O3.2 RP11-774O3.1 |
| chr11_-_72432950 | 0.08 |
ENST00000426523.1
ENST00000429686.1 |
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr10_+_5726764 | 0.08 |
ENST00000328090.5
ENST00000496681.1 |
FAM208B
|
family with sequence similarity 208, member B |
| chr4_+_95917383 | 0.08 |
ENST00000512312.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr16_-_30006922 | 0.08 |
ENST00000564026.1
|
HIRIP3
|
HIRA interacting protein 3 |
| chr7_-_6746474 | 0.07 |
ENST00000394917.3
ENST00000405858.1 ENST00000342651.5 |
ZNF12
|
zinc finger protein 12 |
| chr8_-_23021533 | 0.07 |
ENST00000312584.3
|
TNFRSF10D
|
tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain |
| chr17_+_1646130 | 0.07 |
ENST00000453066.1
ENST00000324015.3 ENST00000450523.2 ENST00000453723.1 ENST00000382061.4 |
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2 |
| chr11_+_393428 | 0.07 |
ENST00000533249.1
ENST00000527442.1 |
PKP3
|
plakophilin 3 |
| chr6_-_101329157 | 0.07 |
ENST00000369143.2
|
ASCC3
|
activating signal cointegrator 1 complex subunit 3 |
| chr8_-_144413981 | 0.07 |
ENST00000522041.1
|
TOP1MT
|
topoisomerase (DNA) I, mitochondrial |
| chr10_-_82049424 | 0.07 |
ENST00000372213.3
|
MAT1A
|
methionine adenosyltransferase I, alpha |
| chr12_-_15942503 | 0.07 |
ENST00000281172.5
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr7_+_102389434 | 0.07 |
ENST00000409231.3
ENST00000418198.1 |
FAM185A
|
family with sequence similarity 185, member A |
| chr3_-_50340996 | 0.07 |
ENST00000266031.4
ENST00000395143.2 ENST00000457214.2 ENST00000447605.2 ENST00000418723.1 ENST00000395144.2 |
HYAL1
|
hyaluronoglucosaminidase 1 |
| chr1_-_114447620 | 0.07 |
ENST00000369569.1
ENST00000432415.1 ENST00000369571.2 |
AP4B1
|
adaptor-related protein complex 4, beta 1 subunit |
| chr19_-_12662314 | 0.07 |
ENST00000339282.7
ENST00000596193.1 |
ZNF564
|
zinc finger protein 564 |
| chr17_+_49230897 | 0.07 |
ENST00000393196.3
ENST00000336097.3 ENST00000480143.1 ENST00000511355.1 ENST00000013034.3 ENST00000393198.3 ENST00000608447.1 ENST00000393193.2 ENST00000376392.6 ENST00000555572.1 |
NME1
NME1-NME2
NME2
|
NME/NM23 nucleoside diphosphate kinase 1 NME1-NME2 readthrough NME/NM23 nucleoside diphosphate kinase 2 |
| chr1_+_116915270 | 0.07 |
ENST00000418797.1
|
ATP1A1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
| chr11_+_64889773 | 0.07 |
ENST00000534078.1
ENST00000526171.1 ENST00000279242.2 ENST00000531705.1 ENST00000533943.1 |
MRPL49
|
mitochondrial ribosomal protein L49 |
| chr1_-_46216286 | 0.07 |
ENST00000396478.3
ENST00000359942.4 |
IPP
|
intracisternal A particle-promoted polypeptide |
| chr4_+_72204755 | 0.07 |
ENST00000512686.1
ENST00000340595.3 |
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr11_+_77899842 | 0.07 |
ENST00000530267.1
|
USP35
|
ubiquitin specific peptidase 35 |
| chr17_-_41277467 | 0.07 |
ENST00000494123.1
ENST00000346315.3 ENST00000309486.4 ENST00000468300.1 ENST00000354071.3 ENST00000352993.3 ENST00000471181.2 |
BRCA1
|
breast cancer 1, early onset |
| chr16_+_718147 | 0.07 |
ENST00000561929.1
|
RHOT2
|
ras homolog family member T2 |
| chr17_+_75450075 | 0.07 |
ENST00000592951.1
|
SEPT9
|
septin 9 |
| chr11_-_18034430 | 0.06 |
ENST00000530613.1
ENST00000532389.1 ENST00000529728.1 ENST00000532265.1 |
SERGEF
|
secretion regulating guanine nucleotide exchange factor |
| chr12_+_108956355 | 0.06 |
ENST00000535729.1
ENST00000431221.2 ENST00000547005.1 ENST00000311893.9 ENST00000392807.4 ENST00000338291.4 ENST00000539593.1 |
ISCU
|
iron-sulfur cluster assembly enzyme |
| chr1_-_114447683 | 0.06 |
ENST00000256658.4
ENST00000369564.1 |
AP4B1
|
adaptor-related protein complex 4, beta 1 subunit |
| chr2_+_108994466 | 0.06 |
ENST00000272452.2
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr19_-_3500635 | 0.06 |
ENST00000250937.3
|
DOHH
|
deoxyhypusine hydroxylase/monooxygenase |
| chr15_+_40226304 | 0.06 |
ENST00000559624.1
ENST00000382727.2 ENST00000263791.5 ENST00000560648.1 |
EIF2AK4
|
eukaryotic translation initiation factor 2 alpha kinase 4 |
| chr15_-_55700457 | 0.06 |
ENST00000442196.3
ENST00000563171.1 ENST00000425574.3 |
CCPG1
|
cell cycle progression 1 |
| chr11_+_60635035 | 0.06 |
ENST00000278853.5
|
ZP1
|
zona pellucida glycoprotein 1 (sperm receptor) |
| chr19_+_45165545 | 0.06 |
ENST00000592472.1
ENST00000587729.1 ENST00000585657.1 ENST00000592789.1 ENST00000591979.1 |
CEACAM19
|
carcinoembryonic antigen-related cell adhesion molecule 19 |
| chr12_-_126467906 | 0.06 |
ENST00000507313.1
ENST00000545784.1 |
LINC00939
|
long intergenic non-protein coding RNA 939 |
| chr10_-_101190202 | 0.06 |
ENST00000543866.1
ENST00000370508.5 |
GOT1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr16_+_32264645 | 0.06 |
ENST00000569631.1
ENST00000354614.3 |
TP53TG3D
|
TP53 target 3D |
| chr19_+_44081344 | 0.06 |
ENST00000599207.1
|
PINLYP
|
phospholipase A2 inhibitor and LY6/PLAUR domain containing |
| chr19_-_10420459 | 0.06 |
ENST00000403352.1
ENST00000403903.3 |
ZGLP1
|
zinc finger, GATA-like protein 1 |
| chr16_+_53483983 | 0.06 |
ENST00000544545.1
|
RBL2
|
retinoblastoma-like 2 (p130) |
| chr11_+_46366918 | 0.06 |
ENST00000528615.1
ENST00000395574.3 |
DGKZ
|
diacylglycerol kinase, zeta |
| chr1_-_46089639 | 0.06 |
ENST00000445048.2
|
CCDC17
|
coiled-coil domain containing 17 |
| chr19_+_49497121 | 0.06 |
ENST00000413176.2
|
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr16_-_75467274 | 0.06 |
ENST00000566254.1
|
CFDP1
|
craniofacial development protein 1 |
| chr22_-_38577731 | 0.06 |
ENST00000335539.3
|
PLA2G6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr3_+_44379611 | 0.06 |
ENST00000383746.3
ENST00000417237.1 |
TCAIM
|
T cell activation inhibitor, mitochondrial |
| chr8_+_22853345 | 0.06 |
ENST00000522948.1
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr5_-_72861175 | 0.06 |
ENST00000504641.1
|
ANKRA2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr15_-_55700522 | 0.06 |
ENST00000564092.1
ENST00000310958.6 |
CCPG1
|
cell cycle progression 1 |
| chr17_-_28661065 | 0.06 |
ENST00000328886.4
ENST00000538566.2 |
TMIGD1
|
transmembrane and immunoglobulin domain containing 1 |
| chr2_-_68384603 | 0.06 |
ENST00000406245.2
ENST00000409164.1 ENST00000295121.6 |
WDR92
|
WD repeat domain 92 |
| chr15_-_29561979 | 0.06 |
ENST00000332303.4
|
NDNL2
|
necdin-like 2 |
| chr12_+_110172572 | 0.05 |
ENST00000358906.3
|
FAM222A
|
family with sequence similarity 222, member A |
| chr1_+_97187318 | 0.05 |
ENST00000609116.1
ENST00000370198.1 ENST00000370197.1 ENST00000426398.2 ENST00000394184.3 |
PTBP2
|
polypyrimidine tract binding protein 2 |
| chr7_+_140396465 | 0.05 |
ENST00000476279.1
ENST00000247866.4 ENST00000461457.1 ENST00000465506.1 ENST00000204307.5 ENST00000464566.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr9_+_131218408 | 0.05 |
ENST00000351030.3
ENST00000604420.1 ENST00000535026.1 ENST00000448249.3 ENST00000393527.3 |
ODF2
|
outer dense fiber of sperm tails 2 |
| chr17_-_14140166 | 0.05 |
ENST00000420162.2
ENST00000431716.2 |
CDRT15
|
CMT1A duplicated region transcript 15 |
| chr2_-_27603582 | 0.05 |
ENST00000323703.6
ENST00000436006.1 |
ZNF513
|
zinc finger protein 513 |
| chr11_+_111783450 | 0.05 |
ENST00000537382.1
|
HSPB2
|
Homo sapiens heat shock 27kDa protein 2 (HSPB2), mRNA. |
| chr14_+_24702073 | 0.05 |
ENST00000399440.2
|
GMPR2
|
guanosine monophosphate reductase 2 |
| chr19_-_11639931 | 0.05 |
ENST00000592312.1
ENST00000590480.1 ENST00000585318.1 ENST00000252440.7 ENST00000417981.2 ENST00000270517.7 |
ECSIT
|
ECSIT signalling integrator |
| chrX_-_84363974 | 0.05 |
ENST00000395409.3
ENST00000332921.5 ENST00000509231.1 |
SATL1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr22_+_39378346 | 0.05 |
ENST00000407298.3
|
APOBEC3B
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3B |
| chr14_-_37641618 | 0.05 |
ENST00000555449.1
|
SLC25A21
|
solute carrier family 25 (mitochondrial oxoadipate carrier), member 21 |
| chr16_-_57837129 | 0.05 |
ENST00000562984.1
ENST00000564891.1 |
KIFC3
|
kinesin family member C3 |
| chr2_+_95873257 | 0.05 |
ENST00000425953.1
|
AC092835.2
|
AC092835.2 |
| chr17_-_72855989 | 0.05 |
ENST00000293190.5
|
GRIN2C
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2C |
| chr19_+_51293672 | 0.05 |
ENST00000270593.1
ENST00000270594.3 |
ACPT
|
acid phosphatase, testicular |
| chr9_+_131218336 | 0.05 |
ENST00000372814.3
|
ODF2
|
outer dense fiber of sperm tails 2 |
| chr19_-_10613421 | 0.05 |
ENST00000393623.2
|
KEAP1
|
kelch-like ECH-associated protein 1 |
| chr3_+_122044084 | 0.05 |
ENST00000264474.3
ENST00000479204.1 |
CSTA
|
cystatin A (stefin A) |
| chr1_-_47697387 | 0.05 |
ENST00000371884.2
|
TAL1
|
T-cell acute lymphocytic leukemia 1 |
| chr3_+_152552685 | 0.05 |
ENST00000305097.3
|
P2RY1
|
purinergic receptor P2Y, G-protein coupled, 1 |
| chr1_-_185286461 | 0.05 |
ENST00000367498.3
|
IVNS1ABP
|
influenza virus NS1A binding protein |
| chr4_+_6784401 | 0.05 |
ENST00000425103.1
ENST00000307659.5 |
KIAA0232
|
KIAA0232 |
| chr7_+_75024903 | 0.05 |
ENST00000323819.3
ENST00000430211.1 |
TRIM73
|
tripartite motif containing 73 |
| chr13_-_48612067 | 0.05 |
ENST00000543413.1
|
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr4_-_7436671 | 0.05 |
ENST00000319098.4
|
PSAPL1
|
prosaposin-like 1 (gene/pseudogene) |
| chr8_+_67025516 | 0.05 |
ENST00000517961.2
|
AC084082.3
|
AC084082.3 |
| chr12_-_56320887 | 0.05 |
ENST00000398213.4
|
WIBG
|
within bgcn homolog (Drosophila) |
| chr1_+_110198689 | 0.05 |
ENST00000369836.4
|
GSTM4
|
glutathione S-transferase mu 4 |
| chr1_-_153588334 | 0.05 |
ENST00000476873.1
|
S100A14
|
S100 calcium binding protein A14 |
| chr2_-_31361543 | 0.05 |
ENST00000349752.5
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr19_+_8509842 | 0.05 |
ENST00000325495.4
ENST00000600092.1 ENST00000594907.1 ENST00000596984.1 ENST00000601645.1 |
HNRNPM
|
heterogeneous nuclear ribonucleoprotein M |
| chr9_-_6007787 | 0.05 |
ENST00000399933.3
ENST00000381461.2 ENST00000513355.2 |
KIAA2026
|
KIAA2026 |
| chr1_+_24646263 | 0.04 |
ENST00000524724.1
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr9_-_132383055 | 0.04 |
ENST00000372478.4
|
C9orf50
|
chromosome 9 open reading frame 50 |
| chr2_+_178257372 | 0.04 |
ENST00000264167.4
ENST00000409888.1 |
AGPS
|
alkylglycerone phosphate synthase |
| chr5_+_170814803 | 0.04 |
ENST00000521672.1
ENST00000351986.6 ENST00000393820.2 ENST00000523622.1 |
NPM1
|
nucleophosmin (nucleolar phosphoprotein B23, numatrin) |
| chr9_-_70488865 | 0.04 |
ENST00000377392.5
|
CBWD5
|
COBW domain containing 5 |
| chr7_+_23145884 | 0.04 |
ENST00000409689.1
ENST00000410047.1 |
KLHL7
|
kelch-like family member 7 |
| chr11_+_67776012 | 0.04 |
ENST00000539229.1
|
ALDH3B1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr3_+_32023232 | 0.04 |
ENST00000360311.4
|
ZNF860
|
zinc finger protein 860 |
| chr18_-_55470320 | 0.04 |
ENST00000536015.1
|
ATP8B1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
| chr19_-_5293243 | 0.04 |
ENST00000591760.1
|
PTPRS
|
protein tyrosine phosphatase, receptor type, S |
| chr1_-_46089718 | 0.04 |
ENST00000421127.2
ENST00000343901.2 ENST00000528266.1 |
CCDC17
|
coiled-coil domain containing 17 |
| chr11_+_65122216 | 0.04 |
ENST00000309880.5
|
TIGD3
|
tigger transposable element derived 3 |
| chr6_-_111804905 | 0.04 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr17_+_78397320 | 0.04 |
ENST00000521634.1
ENST00000572886.1 |
ENDOV
|
endonuclease V |
| chr4_+_25378912 | 0.04 |
ENST00000510092.1
ENST00000505991.1 |
ANAPC4
|
anaphase promoting complex subunit 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TP63
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0071301 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.1 | 0.4 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.4 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.1 | 0.2 | GO:0060266 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.6 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0045575 | basophil activation involved in immune response(GO:0002276) basophil activation(GO:0045575) |
| 0.0 | 0.2 | GO:0044858 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.2 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.2 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.0 | 0.2 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0033025 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.1 | GO:0048597 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.0 | 0.5 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.2 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.1 | GO:0070512 | regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 0.0 | 0.1 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.1 | GO:0010585 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.0 | 0.2 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:0071262 | regulation of eIF2 alpha phosphorylation by amino acid starvation(GO:0060733) regulation of translational initiation in response to starvation(GO:0071262) positive regulation of translational initiation in response to starvation(GO:0071264) |
| 0.0 | 0.1 | GO:0060031 | mediolateral intercalation(GO:0060031) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) negative regulation of fibroblast growth factor production(GO:0090272) |
| 0.0 | 0.1 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.1 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.1 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.1 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 | 0.0 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 0.0 | 0.1 | GO:0031627 | telomeric loop formation(GO:0031627) telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.1 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.0 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 0.1 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.1 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.2 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.0 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.0 | 0.2 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:0002034 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.0 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.0 | 0.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.2 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.1 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.1 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.0 | 0.2 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.1 | 0.2 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
| 0.0 | 0.1 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) |
| 0.0 | 0.1 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.1 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.0 | 0.1 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.0 | 0.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.1 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.0 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.0 | 0.2 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) polyamine binding(GO:0019808) |
| 0.0 | 0.2 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.0 | GO:0031896 | alpha-1A adrenergic receptor binding(GO:0031691) follicle-stimulating hormone receptor binding(GO:0031762) V2 vasopressin receptor binding(GO:0031896) |
| 0.0 | 0.0 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.0 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.1 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.0 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |