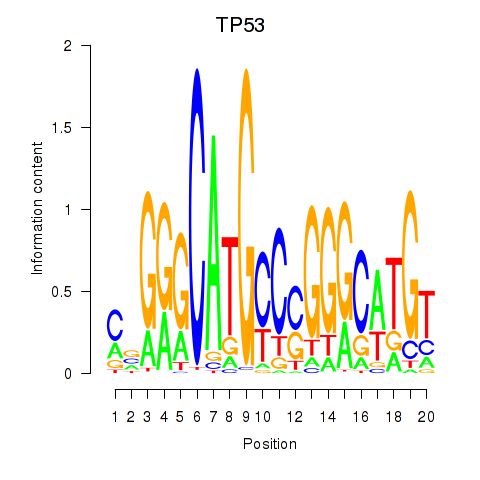
|
chr12_-_53207842
Show fit
|
1.88 |
ENST00000458244.2
|
KRT4
|
keratin 4
|
|
chr10_-_90751038
Show fit
|
1.36 |
ENST00000458159.1
ENST00000415557.1
ENST00000458208.1
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr17_+_76210267
Show fit
|
1.29 |
ENST00000301633.4
ENST00000350051.3
ENST00000374948.2
ENST00000590449.1
|
BIRC5
|
baculoviral IAP repeat containing 5
|
|
chr19_+_28284803
Show fit
|
1.22 |
ENST00000586220.1
ENST00000588784.1
ENST00000591549.1
ENST00000585827.1
ENST00000588636.1
ENST00000587188.1
|
CTC-459F4.3
|
CTC-459F4.3
|
|
chr16_-_75467274
Show fit
|
1.19 |
ENST00000566254.1
|
CFDP1
|
craniofacial development protein 1
|
|
chrX_-_65858865
Show fit
|
1.19 |
ENST00000374719.3
ENST00000450752.1
ENST00000451436.2
|
EDA2R
|
ectodysplasin A2 receptor
|
|
chr5_+_162864575
Show fit
|
1.12 |
ENST00000512163.1
ENST00000393929.1
ENST00000340828.2
ENST00000511683.2
ENST00000510097.1
ENST00000511490.2
ENST00000510664.1
|
CCNG1
|
cyclin G1
|
|
chr9_+_139846708
Show fit
|
1.09 |
ENST00000371633.3
|
LCN12
|
lipocalin 12
|
|
chr19_-_47735918
Show fit
|
1.08 |
ENST00000449228.1
ENST00000300880.7
ENST00000341983.4
|
BBC3
|
BCL2 binding component 3
|
|
chr6_-_138428613
Show fit
|
0.98 |
ENST00000421351.3
|
PERP
|
PERP, TP53 apoptosis effector
|
|
chr15_+_89164560
Show fit
|
0.96 |
ENST00000379231.3
ENST00000559528.1
|
AEN
|
apoptosis enhancing nuclease
|
|
chr15_+_89164520
Show fit
|
0.88 |
ENST00000332810.3
|
AEN
|
apoptosis enhancing nuclease
|
|
chr19_+_15783879
Show fit
|
0.88 |
ENST00000551607.1
|
CYP4F12
|
cytochrome P450, family 4, subfamily F, polypeptide 12
|
|
chrX_-_106959631
Show fit
|
0.85 |
ENST00000486554.1
ENST00000372390.4
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr5_+_27472371
Show fit
|
0.82 |
ENST00000512067.1
ENST00000510165.1
ENST00000505775.1
ENST00000514844.1
ENST00000503107.1
|
LINC01021
|
long intergenic non-protein coding RNA 1021
|
|
chr9_-_95527079
Show fit
|
0.81 |
ENST00000356884.6
ENST00000375512.3
|
BICD2
|
bicaudal D homolog 2 (Drosophila)
|
|
chr2_+_197504278
Show fit
|
0.80 |
ENST00000272831.7
ENST00000389175.4
ENST00000472405.2
ENST00000423093.2
|
CCDC150
|
coiled-coil domain containing 150
|
|
chr20_-_5100591
Show fit
|
0.80 |
ENST00000379143.5
|
PCNA
|
proliferating cell nuclear antigen
|
|
chr1_+_117910047
Show fit
|
0.79 |
ENST00000356554.3
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2
|
|
chr8_+_120428546
Show fit
|
0.79 |
ENST00000259526.3
|
NOV
|
nephroblastoma overexpressed
|
|
chr3_+_184279566
Show fit
|
0.78 |
ENST00000330394.2
|
EPHB3
|
EPH receptor B3
|
|
chr1_+_2477831
Show fit
|
0.77 |
ENST00000606645.1
|
RP3-395M20.12
|
RP3-395M20.12
|
|
chr7_-_21985489
Show fit
|
0.76 |
ENST00000356195.5
ENST00000447180.1
ENST00000373934.4
ENST00000457951.1
|
CDCA7L
|
cell division cycle associated 7-like
|
|
chr7_+_92861653
Show fit
|
0.70 |
ENST00000251739.5
ENST00000305866.5
ENST00000544910.1
ENST00000541136.1
ENST00000458530.1
ENST00000535481.1
ENST00000317751.6
|
CCDC132
|
coiled-coil domain containing 132
|
|
chr12_+_69202795
Show fit
|
0.69 |
ENST00000539479.1
ENST00000393415.3
ENST00000523991.1
ENST00000543323.1
ENST00000393416.2
|
MDM2
|
MDM2 oncogene, E3 ubiquitin protein ligase
|
|
chr13_+_48877895
Show fit
|
0.68 |
ENST00000267163.4
|
RB1
|
retinoblastoma 1
|
|
chr14_+_97263641
Show fit
|
0.68 |
ENST00000216639.3
|
VRK1
|
vaccinia related kinase 1
|
|
chr12_-_126467906
Show fit
|
0.68 |
ENST00000507313.1
ENST00000545784.1
|
LINC00939
|
long intergenic non-protein coding RNA 939
|
|
chr1_-_54519134
Show fit
|
0.67 |
ENST00000371341.1
|
TMEM59
|
transmembrane protein 59
|
|
chr3_+_44379944
Show fit
|
0.66 |
ENST00000396078.3
ENST00000342649.4
|
TCAIM
|
T cell activation inhibitor, mitochondrial
|
|
chr1_-_244615425
Show fit
|
0.62 |
ENST00000366535.3
|
ADSS
|
adenylosuccinate synthase
|
|
chr6_-_101329191
Show fit
|
0.62 |
ENST00000324723.6
ENST00000369162.2
ENST00000522650.1
|
ASCC3
|
activating signal cointegrator 1 complex subunit 3
|
|
chr1_+_25598872
Show fit
|
0.61 |
ENST00000328664.4
|
RHD
|
Rh blood group, D antigen
|
|
chr2_-_157198860
Show fit
|
0.61 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr10_+_96305535
Show fit
|
0.61 |
ENST00000419900.1
ENST00000348459.5
ENST00000394045.1
ENST00000394044.1
ENST00000394036.1
|
HELLS
|
helicase, lymphoid-specific
|
|
chr16_-_14788526
Show fit
|
0.58 |
ENST00000438167.3
|
PLA2G10
|
phospholipase A2, group X
|
|
chr19_-_28284793
Show fit
|
0.58 |
ENST00000590523.1
|
LINC00662
|
long intergenic non-protein coding RNA 662
|
|
chr3_-_46037299
Show fit
|
0.58 |
ENST00000296137.2
|
FYCO1
|
FYVE and coiled-coil domain containing 1
|
|
chr1_-_38412683
Show fit
|
0.57 |
ENST00000373024.3
ENST00000373023.2
|
INPP5B
|
inositol polyphosphate-5-phosphatase, 75kDa
|
|
chr7_-_23145288
Show fit
|
0.56 |
ENST00000419813.1
|
KLHL7-AS1
|
KLHL7 antisense RNA 1 (head to head)
|
|
chr22_+_29168652
Show fit
|
0.56 |
ENST00000249064.4
ENST00000444523.1
ENST00000448492.2
ENST00000421503.2
|
CCDC117
|
coiled-coil domain containing 117
|
|
chr11_+_64073699
Show fit
|
0.56 |
ENST00000405666.1
ENST00000468670.1
|
ESRRA
|
estrogen-related receptor alpha
|
|
chr22_-_24093267
Show fit
|
0.54 |
ENST00000341976.3
|
ZNF70
|
zinc finger protein 70
|
|
chr6_-_30709980
Show fit
|
0.53 |
ENST00000416018.1
ENST00000445853.1
ENST00000413165.1
ENST00000418160.1
|
FLOT1
|
flotillin 1
|
|
chr20_-_23807358
Show fit
|
0.53 |
ENST00000304725.2
|
CST2
|
cystatin SA
|
|
chr19_+_49458107
Show fit
|
0.51 |
ENST00000539787.1
ENST00000345358.7
ENST00000391871.3
ENST00000415969.2
ENST00000354470.3
ENST00000506183.1
ENST00000293288.8
|
BAX
|
BCL2-associated X protein
|
|
chr5_+_72861560
Show fit
|
0.51 |
ENST00000296792.4
ENST00000509005.1
ENST00000543251.1
ENST00000508686.1
ENST00000508491.1
|
UTP15
|
UTP15, U3 small nucleolar ribonucleoprotein, homolog (S. cerevisiae)
|
|
chr1_+_114447763
Show fit
|
0.51 |
ENST00000369563.3
|
DCLRE1B
|
DNA cross-link repair 1B
|
|
chr17_-_74528128
Show fit
|
0.50 |
ENST00000590175.1
|
CYGB
|
cytoglobin
|
|
chr11_+_827553
Show fit
|
0.50 |
ENST00000528542.2
ENST00000450448.1
|
EFCAB4A
|
EF-hand calcium binding domain 4A
|
|
chr9_+_139847347
Show fit
|
0.49 |
ENST00000371632.3
|
LCN12
|
lipocalin 12
|
|
chr2_+_11295624
Show fit
|
0.48 |
ENST00000402361.1
ENST00000428481.1
|
PQLC3
|
PQ loop repeat containing 3
|
|
chr14_+_64971292
Show fit
|
0.46 |
ENST00000358738.3
ENST00000394712.2
|
ZBTB1
|
zinc finger and BTB domain containing 1
|
|
chr11_+_827248
Show fit
|
0.46 |
ENST00000527089.1
ENST00000530183.1
|
EFCAB4A
|
EF-hand calcium binding domain 4A
|
|
chr3_+_44379611
Show fit
|
0.46 |
ENST00000383746.3
ENST00000417237.1
|
TCAIM
|
T cell activation inhibitor, mitochondrial
|
|
chr18_-_28622699
Show fit
|
0.46 |
ENST00000360428.4
|
DSC3
|
desmocollin 3
|
|
chr19_-_6481776
Show fit
|
0.46 |
ENST00000543576.1
ENST00000590173.1
ENST00000381480.2
|
DENND1C
|
DENN/MADD domain containing 1C
|
|
chr1_-_54519067
Show fit
|
0.45 |
ENST00000452421.1
ENST00000420738.1
ENST00000234831.5
ENST00000440019.1
|
TMEM59
|
transmembrane protein 59
|
|
chr3_-_113464906
Show fit
|
0.45 |
ENST00000477813.1
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr2_+_11295498
Show fit
|
0.44 |
ENST00000295083.3
ENST00000441908.2
|
PQLC3
|
PQ loop repeat containing 3
|
|
chrX_+_40440146
Show fit
|
0.44 |
ENST00000535539.1
ENST00000378438.4
ENST00000436783.1
ENST00000544975.1
ENST00000535777.1
ENST00000447485.1
ENST00000423649.1
|
ATP6AP2
|
ATPase, H+ transporting, lysosomal accessory protein 2
|
|
chr15_-_56035177
Show fit
|
0.44 |
ENST00000389286.4
ENST00000561292.1
|
PRTG
|
protogenin
|
|
chr18_-_28622774
Show fit
|
0.43 |
ENST00000434452.1
|
DSC3
|
desmocollin 3
|
|
chr10_+_88854926
Show fit
|
0.43 |
ENST00000298784.1
ENST00000298786.4
|
FAM35A
|
family with sequence similarity 35, member A
|
|
chr1_+_204494618
Show fit
|
0.43 |
ENST00000367180.1
ENST00000391947.2
|
MDM4
|
Mdm4 p53 binding protein homolog (mouse)
|
|
chr2_+_169312725
Show fit
|
0.43 |
ENST00000392687.4
|
CERS6
|
ceramide synthase 6
|
|
chr17_-_61523622
Show fit
|
0.42 |
ENST00000448884.2
ENST00000582297.1
ENST00000582034.1
ENST00000578072.1
ENST00000360793.3
|
CYB561
|
cytochrome b561
|
|
chr14_+_36295638
Show fit
|
0.42 |
ENST00000543183.1
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like
|
|
chr14_-_102553371
Show fit
|
0.42 |
ENST00000553585.1
ENST00000216281.8
|
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1
|
|
chr2_+_85661918
Show fit
|
0.42 |
ENST00000340326.2
|
SH2D6
|
SH2 domain containing 6
|
|
chr16_-_4303767
Show fit
|
0.42 |
ENST00000573268.1
ENST00000573042.1
|
RP11-95P2.1
|
RP11-95P2.1
|
|
chr10_+_64564469
Show fit
|
0.42 |
ENST00000373783.1
|
ADO
|
2-aminoethanethiol (cysteamine) dioxygenase
|
|
chr19_+_13988061
Show fit
|
0.41 |
ENST00000339133.5
ENST00000397555.2
|
NANOS3
|
nanos homolog 3 (Drosophila)
|
|
chr16_+_57653854
Show fit
|
0.41 |
ENST00000568908.1
ENST00000568909.1
ENST00000566778.1
ENST00000561988.1
|
GPR56
|
G protein-coupled receptor 56
|
|
chr7_-_56101826
Show fit
|
0.41 |
ENST00000421626.1
|
PSPH
|
phosphoserine phosphatase
|
|
chr12_+_69202975
Show fit
|
0.39 |
ENST00000544561.1
ENST00000393410.1
ENST00000299252.4
ENST00000360430.2
ENST00000517852.1
ENST00000545204.1
ENST00000393413.3
ENST00000350057.5
ENST00000348801.2
ENST00000478070.1
|
MDM2
|
MDM2 oncogene, E3 ubiquitin protein ligase
|
|
chr1_-_213189168
Show fit
|
0.39 |
ENST00000366962.3
ENST00000360506.2
|
ANGEL2
|
angel homolog 2 (Drosophila)
|
|
chr17_-_5095126
Show fit
|
0.38 |
ENST00000576772.1
ENST00000575779.1
|
ZNF594
|
zinc finger protein 594
|
|
chr9_+_104161123
Show fit
|
0.38 |
ENST00000374861.3
ENST00000339664.2
ENST00000259395.4
|
ZNF189
|
zinc finger protein 189
|
|
chr1_+_203830703
Show fit
|
0.37 |
ENST00000414487.2
|
SNRPE
|
small nuclear ribonucleoprotein polypeptide E
|
|
chr6_-_109804412
Show fit
|
0.37 |
ENST00000230122.3
|
ZBTB24
|
zinc finger and BTB domain containing 24
|
|
chr12_-_59314246
Show fit
|
0.37 |
ENST00000320743.3
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr20_-_1373606
Show fit
|
0.37 |
ENST00000381715.1
ENST00000439640.2
ENST00000381719.3
|
FKBP1A
|
FK506 binding protein 1A, 12kDa
|
|
chr15_-_63448973
Show fit
|
0.37 |
ENST00000462430.1
|
RPS27L
|
ribosomal protein S27-like
|
|
chr6_-_101329157
Show fit
|
0.36 |
ENST00000369143.2
|
ASCC3
|
activating signal cointegrator 1 complex subunit 3
|
|
chr4_+_26859300
Show fit
|
0.36 |
ENST00000494628.2
|
STIM2
|
stromal interaction molecule 2
|
|
chr18_+_44497455
Show fit
|
0.36 |
ENST00000592005.1
|
KATNAL2
|
katanin p60 subunit A-like 2
|
|
chr17_-_61523535
Show fit
|
0.36 |
ENST00000584031.1
ENST00000392976.1
|
CYB561
|
cytochrome b561
|
|
chr9_-_115774453
Show fit
|
0.36 |
ENST00000427548.1
|
ZNF883
|
zinc finger protein 883
|
|
chr22_+_42949925
Show fit
|
0.34 |
ENST00000327678.5
ENST00000340239.4
ENST00000407614.4
ENST00000335879.5
|
SERHL2
|
serine hydrolase-like 2
|
|
chr20_+_57467204
Show fit
|
0.34 |
ENST00000603546.1
|
GNAS
|
GNAS complex locus
|
|
chr3_-_44519131
Show fit
|
0.33 |
ENST00000425708.2
ENST00000396077.2
|
ZNF445
|
zinc finger protein 445
|
|
chr8_+_95732095
Show fit
|
0.33 |
ENST00000414645.2
|
DPY19L4
|
dpy-19-like 4 (C. elegans)
|
|
chr7_-_102789629
Show fit
|
0.33 |
ENST00000417955.1
ENST00000341533.4
ENST00000425379.1
|
NAPEPLD
|
N-acyl phosphatidylethanolamine phospholipase D
|
|
chr12_+_99038998
Show fit
|
0.33 |
ENST00000359972.2
ENST00000357310.1
ENST00000339433.3
ENST00000333991.1
|
APAF1
|
apoptotic peptidase activating factor 1
|
|
chr20_-_43589109
Show fit
|
0.33 |
ENST00000372813.3
|
TOMM34
|
translocase of outer mitochondrial membrane 34
|
|
chr4_+_6784401
Show fit
|
0.33 |
ENST00000425103.1
ENST00000307659.5
|
KIAA0232
|
KIAA0232
|
|
chr17_-_4900873
Show fit
|
0.32 |
ENST00000355025.3
ENST00000575780.1
ENST00000396829.2
|
INCA1
|
inhibitor of CDK, cyclin A1 interacting protein 1
|
|
chr8_-_23021533
Show fit
|
0.32 |
ENST00000312584.3
|
TNFRSF10D
|
tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain
|
|
chr4_-_187644930
Show fit
|
0.32 |
ENST00000441802.2
|
FAT1
|
FAT atypical cadherin 1
|
|
chr20_-_1373682
Show fit
|
0.31 |
ENST00000381724.3
|
FKBP1A
|
FK506 binding protein 1A, 12kDa
|
|
chr13_-_50367057
Show fit
|
0.31 |
ENST00000261667.3
|
KPNA3
|
karyopherin alpha 3 (importin alpha 4)
|
|
chr4_+_41937131
Show fit
|
0.31 |
ENST00000504986.1
ENST00000508448.1
ENST00000513702.1
ENST00000325094.5
|
TMEM33
|
transmembrane protein 33
|
|
chr5_-_72861175
Show fit
|
0.30 |
ENST00000504641.1
|
ANKRA2
|
ankyrin repeat, family A (RFXANK-like), 2
|
|
chr6_+_24403144
Show fit
|
0.30 |
ENST00000274747.7
ENST00000543597.1
ENST00000535061.1
ENST00000378353.1
ENST00000378386.3
ENST00000443868.2
|
MRS2
|
MRS2 magnesium transporter
|
|
chr9_+_80912059
Show fit
|
0.29 |
ENST00000347159.2
ENST00000376588.3
|
PSAT1
|
phosphoserine aminotransferase 1
|
|
chr10_-_88854518
Show fit
|
0.29 |
ENST00000277865.4
|
GLUD1
|
glutamate dehydrogenase 1
|
|
chr9_-_104160872
Show fit
|
0.29 |
ENST00000539624.1
ENST00000374865.4
|
MRPL50
|
mitochondrial ribosomal protein L50
|
|
chr9_-_140115775
Show fit
|
0.29 |
ENST00000391553.1
ENST00000392827.1
|
RNF208
|
ring finger protein 208
|
|
chr7_-_100239132
Show fit
|
0.29 |
ENST00000223051.3
ENST00000431692.1
|
TFR2
|
transferrin receptor 2
|
|
chr14_-_36789783
Show fit
|
0.28 |
ENST00000605579.1
ENST00000604336.1
ENST00000359527.7
ENST00000603139.1
ENST00000318473.7
|
MBIP
|
MAP3K12 binding inhibitory protein 1
|
|
chr10_-_105212141
Show fit
|
0.28 |
ENST00000369788.3
|
CALHM2
|
calcium homeostasis modulator 2
|
|
chr2_+_169312350
Show fit
|
0.28 |
ENST00000305747.6
|
CERS6
|
ceramide synthase 6
|
|
chr6_+_33378517
Show fit
|
0.28 |
ENST00000428274.1
|
PHF1
|
PHD finger protein 1
|
|
chr16_+_57653989
Show fit
|
0.28 |
ENST00000567835.1
ENST00000569372.1
ENST00000563548.1
ENST00000562003.1
|
GPR56
|
G protein-coupled receptor 56
|
|
chr1_-_205744574
Show fit
|
0.27 |
ENST00000367139.3
ENST00000235932.4
ENST00000437324.2
ENST00000414729.1
|
RAB7L1
|
RAB7, member RAS oncogene family-like 1
|
|
chr1_-_1356719
Show fit
|
0.27 |
ENST00000520296.1
|
ANKRD65
|
ankyrin repeat domain 65
|
|
chr1_-_114447620
Show fit
|
0.27 |
ENST00000369569.1
ENST00000432415.1
ENST00000369571.2
|
AP4B1
|
adaptor-related protein complex 4, beta 1 subunit
|
|
chr11_-_44972476
Show fit
|
0.27 |
ENST00000527685.1
ENST00000308212.5
|
TP53I11
|
tumor protein p53 inducible protein 11
|
|
chr10_+_96305610
Show fit
|
0.27 |
ENST00000371332.4
ENST00000239026.6
|
HELLS
|
helicase, lymphoid-specific
|
|
chr12_-_14721283
Show fit
|
0.27 |
ENST00000240617.5
|
PLBD1
|
phospholipase B domain containing 1
|
|
chrX_+_134478706
Show fit
|
0.27 |
ENST00000370761.3
ENST00000339249.4
ENST00000370760.3
|
ZNF449
|
zinc finger protein 449
|
|
chr17_+_14204389
Show fit
|
0.26 |
ENST00000360954.2
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1
|
|
chr8_+_22429205
Show fit
|
0.26 |
ENST00000520207.1
|
SORBS3
|
sorbin and SH3 domain containing 3
|
|
chr7_+_23145884
Show fit
|
0.26 |
ENST00000409689.1
ENST00000410047.1
|
KLHL7
|
kelch-like family member 7
|
|
chr14_+_64970662
Show fit
|
0.26 |
ENST00000556965.1
ENST00000554015.1
|
ZBTB1
|
zinc finger and BTB domain containing 1
|
|
chr1_-_1356628
Show fit
|
0.25 |
ENST00000442470.1
ENST00000537107.1
|
ANKRD65
|
ankyrin repeat domain 65
|
|
chr22_-_28392227
Show fit
|
0.25 |
ENST00000431039.1
|
TTC28
|
tetratricopeptide repeat domain 28
|
|
chr2_-_167232484
Show fit
|
0.24 |
ENST00000375387.4
ENST00000303354.6
ENST00000409672.1
|
SCN9A
|
sodium channel, voltage-gated, type IX, alpha subunit
|
|
chr3_-_52804872
Show fit
|
0.24 |
ENST00000535191.1
ENST00000461689.1
ENST00000383721.4
ENST00000233027.5
|
NEK4
|
NIMA-related kinase 4
|
|
chr19_+_55999916
Show fit
|
0.24 |
ENST00000587166.1
ENST00000389623.6
|
SSC5D
|
scavenger receptor cysteine rich domain containing (5 domains)
|
|
chr1_-_153588334
Show fit
|
0.24 |
ENST00000476873.1
|
S100A14
|
S100 calcium binding protein A14
|
|
chr11_-_44972390
Show fit
|
0.24 |
ENST00000395648.3
ENST00000531928.2
|
TP53I11
|
tumor protein p53 inducible protein 11
|
|
chr16_+_77756399
Show fit
|
0.24 |
ENST00000564085.1
ENST00000268533.5
ENST00000568787.1
ENST00000437314.3
ENST00000563839.1
|
NUDT7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7
|
|
chr19_-_2456922
Show fit
|
0.24 |
ENST00000582871.1
ENST00000325327.3
|
LMNB2
|
lamin B2
|
|
chr7_+_75024903
Show fit
|
0.23 |
ENST00000323819.3
ENST00000430211.1
|
TRIM73
|
tripartite motif containing 73
|
|
chr11_+_1855645
Show fit
|
0.23 |
ENST00000381968.3
ENST00000381978.3
|
SYT8
|
synaptotagmin VIII
|
|
chr8_-_12612962
Show fit
|
0.23 |
ENST00000398246.3
|
LONRF1
|
LON peptidase N-terminal domain and ring finger 1
|
|
chrX_-_70474499
Show fit
|
0.23 |
ENST00000353904.2
|
ZMYM3
|
zinc finger, MYM-type 3
|
|
chr5_-_72861484
Show fit
|
0.23 |
ENST00000296785.3
|
ANKRA2
|
ankyrin repeat, family A (RFXANK-like), 2
|
|
chr17_-_72855989
Show fit
|
0.23 |
ENST00000293190.5
|
GRIN2C
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2C
|
|
chr1_-_54411240
Show fit
|
0.22 |
ENST00000371378.2
|
HSPB11
|
heat shock protein family B (small), member 11
|
|
chr1_+_17634689
Show fit
|
0.22 |
ENST00000375453.1
ENST00000375448.4
|
PADI4
|
peptidyl arginine deiminase, type IV
|
|
chr2_-_75788428
Show fit
|
0.22 |
ENST00000432649.1
|
EVA1A
|
eva-1 homolog A (C. elegans)
|
|
chr1_-_26197744
Show fit
|
0.22 |
ENST00000374296.3
|
PAQR7
|
progestin and adipoQ receptor family member VII
|
|
chrX_-_70474377
Show fit
|
0.22 |
ENST00000373978.1
ENST00000373981.1
|
ZMYM3
|
zinc finger, MYM-type 3
|
|
chr11_-_129062093
Show fit
|
0.22 |
ENST00000310343.9
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr1_-_25747283
Show fit
|
0.22 |
ENST00000346452.4
ENST00000340849.4
ENST00000349438.4
ENST00000294413.7
ENST00000413854.1
ENST00000455194.1
ENST00000243186.6
ENST00000425135.1
|
RHCE
|
Rh blood group, CcEe antigens
|
|
chr1_-_243418650
Show fit
|
0.21 |
ENST00000522995.1
|
CEP170
|
centrosomal protein 170kDa
|
|
chrX_+_152086373
Show fit
|
0.21 |
ENST00000318529.8
|
ZNF185
|
zinc finger protein 185 (LIM domain)
|
|
chr4_+_671711
Show fit
|
0.21 |
ENST00000400159.2
|
MYL5
|
myosin, light chain 5, regulatory
|
|
chr8_-_22926623
Show fit
|
0.21 |
ENST00000276431.4
|
TNFRSF10B
|
tumor necrosis factor receptor superfamily, member 10b
|
|
chr11_-_44972418
Show fit
|
0.21 |
ENST00000525680.1
ENST00000528290.1
ENST00000530035.1
|
TP53I11
|
tumor protein p53 inducible protein 11
|
|
chr14_+_24702127
Show fit
|
0.20 |
ENST00000557854.1
ENST00000348719.7
ENST00000559104.1
ENST00000456667.3
|
GMPR2
|
guanosine monophosphate reductase 2
|
|
chr11_-_77123065
Show fit
|
0.20 |
ENST00000530617.1
|
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1
|
|
chr22_-_39715600
Show fit
|
0.20 |
ENST00000427905.1
ENST00000402527.1
ENST00000216146.4
|
RPL3
|
ribosomal protein L3
|
|
chr12_+_44229846
Show fit
|
0.20 |
ENST00000551577.1
ENST00000266534.3
|
TMEM117
|
transmembrane protein 117
|
|
chr17_+_76210367
Show fit
|
0.20 |
ENST00000592734.1
ENST00000587746.1
|
BIRC5
|
baculoviral IAP repeat containing 5
|
|
chr1_+_27153173
Show fit
|
0.20 |
ENST00000374142.4
|
ZDHHC18
|
zinc finger, DHHC-type containing 18
|
|
chr19_+_489140
Show fit
|
0.20 |
ENST00000587541.1
|
MADCAM1
|
mucosal vascular addressin cell adhesion molecule 1
|
|
chr17_+_20483037
Show fit
|
0.20 |
ENST00000399044.1
|
CDRT15L2
|
CMT1A duplicated region transcript 15-like 2
|
|
chr1_+_54411715
Show fit
|
0.20 |
ENST00000371370.3
ENST00000371368.1
|
LRRC42
|
leucine rich repeat containing 42
|
|
chr19_+_51293672
Show fit
|
0.20 |
ENST00000270593.1
ENST00000270594.3
|
ACPT
|
acid phosphatase, testicular
|
|
chrX_-_70473957
Show fit
|
0.20 |
ENST00000373984.3
ENST00000314425.5
ENST00000373982.1
|
ZMYM3
|
zinc finger, MYM-type 3
|
|
chr8_-_12051576
Show fit
|
0.20 |
ENST00000524571.2
ENST00000533852.2
ENST00000533513.1
ENST00000448228.2
ENST00000534520.1
ENST00000321602.8
|
FAM86B1
|
family with sequence similarity 86, member B1
|
|
chr10_+_11784360
Show fit
|
0.19 |
ENST00000379215.4
ENST00000420401.1
|
ECHDC3
|
enoyl CoA hydratase domain containing 3
|
|
chr3_+_149530469
Show fit
|
0.19 |
ENST00000392894.3
ENST00000344229.3
|
RNF13
|
ring finger protein 13
|
|
chr8_-_144413981
Show fit
|
0.19 |
ENST00000522041.1
|
TOP1MT
|
topoisomerase (DNA) I, mitochondrial
|
|
chr16_-_29874462
Show fit
|
0.19 |
ENST00000566113.1
ENST00000569956.1
ENST00000570016.1
|
CDIPT
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase
|
|
chr17_-_37309480
Show fit
|
0.19 |
ENST00000539608.1
|
PLXDC1
|
plexin domain containing 1
|
|
chr22_-_38484922
Show fit
|
0.19 |
ENST00000428572.1
|
BAIAP2L2
|
BAI1-associated protein 2-like 2
|
|
chr8_-_69243711
Show fit
|
0.19 |
ENST00000512294.3
|
RP11-664D7.4
|
HCG1787533; Uncharacterized protein
|
|
chr17_+_46918925
Show fit
|
0.19 |
ENST00000502761.1
|
CALCOCO2
|
calcium binding and coiled-coil domain 2
|
|
chr3_-_100120223
Show fit
|
0.18 |
ENST00000284320.5
|
TOMM70A
|
translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae)
|
|
chr11_+_67171358
Show fit
|
0.18 |
ENST00000526387.1
|
TBC1D10C
|
TBC1 domain family, member 10C
|
|
chr2_-_85555086
Show fit
|
0.18 |
ENST00000444342.2
ENST00000409232.3
ENST00000409015.1
|
TGOLN2
|
trans-golgi network protein 2
|
|
chr7_+_23145366
Show fit
|
0.18 |
ENST00000339077.5
ENST00000322275.5
ENST00000539124.1
ENST00000542558.1
|
KLHL7
|
kelch-like family member 7
|
|
chr1_-_202936394
Show fit
|
0.18 |
ENST00000367249.4
|
CYB5R1
|
cytochrome b5 reductase 1
|
|
chr10_+_126490354
Show fit
|
0.18 |
ENST00000298492.5
|
FAM175B
|
family with sequence similarity 175, member B
|
|
chr14_-_36789865
Show fit
|
0.18 |
ENST00000416007.4
|
MBIP
|
MAP3K12 binding inhibitory protein 1
|
|
chr1_+_54411995
Show fit
|
0.18 |
ENST00000319223.4
ENST00000444987.1
|
LRRC42
|
leucine rich repeat containing 42
|
|
chr11_+_706595
Show fit
|
0.17 |
ENST00000531348.1
ENST00000530636.1
|
EPS8L2
|
EPS8-like 2
|
|
chr17_-_14140166
Show fit
|
0.17 |
ENST00000420162.2
ENST00000431716.2
|
CDRT15
|
CMT1A duplicated region transcript 15
|
|
chr16_-_29874211
Show fit
|
0.17 |
ENST00000563415.1
|
CDIPT
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase
|
|
chr1_-_205744205
Show fit
|
0.17 |
ENST00000446390.2
|
RAB7L1
|
RAB7, member RAS oncogene family-like 1
|
|
chr7_+_116165754
Show fit
|
0.16 |
ENST00000405348.1
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr14_-_50999307
Show fit
|
0.16 |
ENST00000013125.4
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5
|
|
chr12_-_122711968
Show fit
|
0.16 |
ENST00000485724.1
|
DIABLO
|
diablo, IAP-binding mitochondrial protein
|
|
chr2_-_242556900
Show fit
|
0.16 |
ENST00000402545.1
ENST00000402136.1
|
THAP4
|
THAP domain containing 4
|
|
chr1_-_54518865
Show fit
|
0.16 |
ENST00000371337.3
|
TMEM59
|
transmembrane protein 59
|
|
chr17_+_80332153
Show fit
|
0.16 |
ENST00000313135.2
|
UTS2R
|
urotensin 2 receptor
|
|
chr10_-_105212059
Show fit
|
0.15 |
ENST00000260743.5
|
CALHM2
|
calcium homeostasis modulator 2
|
|
chr5_+_141348598
Show fit
|
0.15 |
ENST00000394520.2
ENST00000347642.3
|
RNF14
|
ring finger protein 14
|
|
chr8_-_22926526
Show fit
|
0.15 |
ENST00000347739.3
ENST00000542226.1
|
TNFRSF10B
|
tumor necrosis factor receptor superfamily, member 10b
|
|
chr11_-_68611721
Show fit
|
0.15 |
ENST00000561996.1
|
CPT1A
|
carnitine palmitoyltransferase 1A (liver)
|
|
chr6_-_111804905
Show fit
|
0.15 |
ENST00000358835.3
ENST00000435970.1
|
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit
|
|
chr11_+_119039414
Show fit
|
0.15 |
ENST00000409991.1
ENST00000292199.2
ENST00000409265.4
ENST00000409109.1
|
NLRX1
|
NLR family member X1
|
|
chr1_-_114447683
Show fit
|
0.14 |
ENST00000256658.4
ENST00000369564.1
|
AP4B1
|
adaptor-related protein complex 4, beta 1 subunit
|
|
chr5_+_64920543
Show fit
|
0.14 |
ENST00000399438.3
ENST00000510585.2
|
TRAPPC13
CTC-534A2.2
|
trafficking protein particle complex 13
CDNA FLJ26957 fis, clone SLV00486; Uncharacterized protein
|
|
chr14_-_31889782
Show fit
|
0.14 |
ENST00000543095.2
|
HEATR5A
|
HEAT repeat containing 5A
|