Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for TLX2
Z-value: 1.18
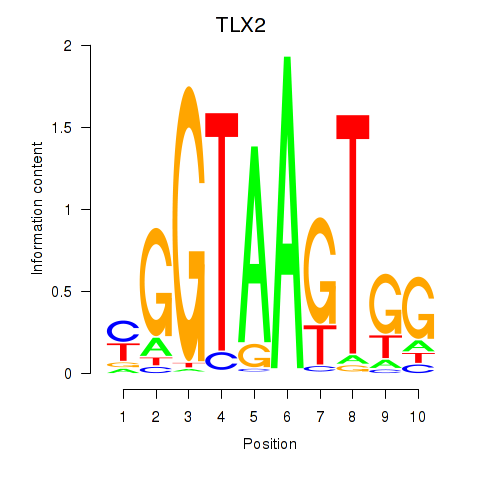
Transcription factors associated with TLX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TLX2
|
ENSG00000115297.9 | T cell leukemia homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TLX2 | hg19_v2_chr2_+_74741569_74741620 | -0.78 | 2.2e-01 | Click! |
Activity profile of TLX2 motif
Sorted Z-values of TLX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_90712520 | 1.21 |
ENST00000224784.6
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr20_+_3024266 | 0.96 |
ENST00000245983.2
ENST00000359100.2 ENST00000359987.1 |
GNRH2
|
gonadotropin-releasing hormone 2 |
| chr3_+_186330712 | 0.95 |
ENST00000411641.2
ENST00000273784.5 |
AHSG
|
alpha-2-HS-glycoprotein |
| chr10_+_115312766 | 0.82 |
ENST00000351270.3
|
HABP2
|
hyaluronan binding protein 2 |
| chr2_-_190445499 | 0.69 |
ENST00000261024.2
|
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr20_-_25038804 | 0.68 |
ENST00000323482.4
|
ACSS1
|
acyl-CoA synthetase short-chain family member 1 |
| chr15_+_82555125 | 0.67 |
ENST00000566205.1
ENST00000339465.5 ENST00000569120.1 ENST00000566861.1 |
FAM154B
|
family with sequence similarity 154, member B |
| chr4_-_105416039 | 0.61 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr10_+_88718314 | 0.60 |
ENST00000348795.4
|
SNCG
|
synuclein, gamma (breast cancer-specific protein 1) |
| chr6_-_138428613 | 0.58 |
ENST00000421351.3
|
PERP
|
PERP, TP53 apoptosis effector |
| chr13_+_113777105 | 0.57 |
ENST00000409306.1
ENST00000375551.3 ENST00000375559.3 |
F10
|
coagulation factor X |
| chr12_-_89918982 | 0.57 |
ENST00000549504.1
|
POC1B
|
POC1 centriolar protein B |
| chr12_-_89918522 | 0.55 |
ENST00000529983.2
|
GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) |
| chr8_+_98900132 | 0.55 |
ENST00000520016.1
|
MATN2
|
matrilin 2 |
| chr7_-_99573677 | 0.53 |
ENST00000292401.4
|
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding |
| chr6_-_41006928 | 0.52 |
ENST00000244565.3
|
UNC5CL
|
unc-5 homolog C (C. elegans)-like |
| chr8_+_120428546 | 0.52 |
ENST00000259526.3
|
NOV
|
nephroblastoma overexpressed |
| chr2_+_128180842 | 0.51 |
ENST00000402125.2
|
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr20_-_52612705 | 0.50 |
ENST00000434986.2
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr3_+_118892362 | 0.50 |
ENST00000497685.1
ENST00000264234.3 |
UPK1B
|
uroplakin 1B |
| chr4_+_165675197 | 0.49 |
ENST00000515485.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr10_+_103348031 | 0.47 |
ENST00000370151.4
ENST00000370147.1 ENST00000370148.2 |
DPCD
|
deleted in primary ciliary dyskinesia homolog (mouse) |
| chr20_-_52612468 | 0.46 |
ENST00000422805.1
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr10_+_115312825 | 0.44 |
ENST00000537906.1
ENST00000541666.1 |
HABP2
|
hyaluronan binding protein 2 |
| chr19_+_16435625 | 0.43 |
ENST00000248071.5
ENST00000592003.1 |
KLF2
|
Kruppel-like factor 2 |
| chr3_+_30647994 | 0.43 |
ENST00000295754.5
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa) |
| chr12_-_99038732 | 0.42 |
ENST00000393042.3
ENST00000420861.1 ENST00000299157.4 ENST00000342502.2 |
IKBIP
|
IKBKB interacting protein |
| chr10_+_88718397 | 0.42 |
ENST00000372017.3
|
SNCG
|
synuclein, gamma (breast cancer-specific protein 1) |
| chr15_-_34502197 | 0.41 |
ENST00000557877.1
|
KATNBL1
|
katanin p80 subunit B-like 1 |
| chr19_-_38878632 | 0.41 |
ENST00000586599.1
ENST00000334928.6 ENST00000587676.1 |
GGN
|
gametogenetin |
| chr3_+_69134124 | 0.41 |
ENST00000478935.1
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr1_-_156399184 | 0.40 |
ENST00000368243.1
ENST00000357975.4 ENST00000310027.5 ENST00000400991.2 |
C1orf61
|
chromosome 1 open reading frame 61 |
| chr15_-_31523036 | 0.40 |
ENST00000559094.1
ENST00000558388.2 |
RP11-16E12.2
|
RP11-16E12.2 |
| chr12_+_65672702 | 0.39 |
ENST00000538045.1
ENST00000535239.1 |
MSRB3
|
methionine sulfoxide reductase B3 |
| chr15_-_34502278 | 0.39 |
ENST00000559515.1
ENST00000256544.3 ENST00000560108.1 ENST00000559462.1 |
KATNBL1
|
katanin p80 subunit B-like 1 |
| chr19_+_15752088 | 0.39 |
ENST00000585846.1
|
CYP4F3
|
cytochrome P450, family 4, subfamily F, polypeptide 3 |
| chr1_+_62417957 | 0.38 |
ENST00000307297.7
ENST00000543708.1 |
INADL
|
InaD-like (Drosophila) |
| chr8_-_27941380 | 0.38 |
ENST00000413272.2
ENST00000341513.6 |
NUGGC
|
nuclear GTPase, germinal center associated |
| chr4_-_84205905 | 0.37 |
ENST00000311461.7
ENST00000311469.4 ENST00000439031.2 |
COQ2
|
coenzyme Q2 4-hydroxybenzoate polyprenyltransferase |
| chr10_-_120938303 | 0.37 |
ENST00000356951.3
ENST00000298510.2 |
PRDX3
|
peroxiredoxin 3 |
| chr17_-_46667628 | 0.37 |
ENST00000498678.1
|
HOXB3
|
homeobox B3 |
| chr9_-_112970436 | 0.37 |
ENST00000400613.4
|
C9orf152
|
chromosome 9 open reading frame 152 |
| chr12_+_65672423 | 0.37 |
ENST00000355192.3
ENST00000308259.5 ENST00000540804.1 ENST00000535664.1 ENST00000541189.1 |
MSRB3
|
methionine sulfoxide reductase B3 |
| chr12_+_50479101 | 0.36 |
ENST00000551966.1
|
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr22_+_50925213 | 0.35 |
ENST00000395733.3
ENST00000216075.6 ENST00000395732.3 |
MIOX
|
myo-inositol oxygenase |
| chr17_-_46667594 | 0.35 |
ENST00000476342.1
ENST00000460160.1 ENST00000472863.1 |
HOXB3
|
homeobox B3 |
| chr3_+_118892411 | 0.35 |
ENST00000479520.1
ENST00000494855.1 |
UPK1B
|
uroplakin 1B |
| chr4_+_17812525 | 0.35 |
ENST00000251496.2
|
NCAPG
|
non-SMC condensin I complex, subunit G |
| chr4_-_68411275 | 0.35 |
ENST00000273853.6
|
CENPC
|
centromere protein C |
| chr12_+_6602517 | 0.35 |
ENST00000315579.5
ENST00000539714.1 |
NCAPD2
|
non-SMC condensin I complex, subunit D2 |
| chrX_-_45629661 | 0.34 |
ENST00000602507.1
ENST00000602461.1 |
RP6-99M1.2
|
RP6-99M1.2 |
| chr1_+_53480598 | 0.34 |
ENST00000430330.2
ENST00000408941.3 ENST00000478274.2 ENST00000484100.1 ENST00000435345.2 ENST00000488965.1 |
SCP2
|
sterol carrier protein 2 |
| chr8_+_38758737 | 0.34 |
ENST00000521746.1
ENST00000420274.1 |
PLEKHA2
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 |
| chr5_+_102455968 | 0.34 |
ENST00000358359.3
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr8_+_74903580 | 0.33 |
ENST00000284818.2
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr16_-_3149278 | 0.33 |
ENST00000575108.1
ENST00000576483.1 ENST00000538082.2 ENST00000576985.1 |
ZSCAN10
|
zinc finger and SCAN domain containing 10 |
| chr16_+_53412368 | 0.33 |
ENST00000565189.1
|
RP11-44F14.2
|
RP11-44F14.2 |
| chr19_-_51920952 | 0.33 |
ENST00000356298.5
ENST00000339313.5 ENST00000529627.1 ENST00000439889.2 ENST00000353836.5 ENST00000432469.2 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chr16_+_53483983 | 0.33 |
ENST00000544545.1
|
RBL2
|
retinoblastoma-like 2 (p130) |
| chr6_+_43968306 | 0.32 |
ENST00000442114.2
ENST00000336600.5 ENST00000439969.2 |
C6orf223
|
chromosome 6 open reading frame 223 |
| chr9_+_139847347 | 0.32 |
ENST00000371632.3
|
LCN12
|
lipocalin 12 |
| chr2_-_204400013 | 0.32 |
ENST00000374489.2
ENST00000374488.2 ENST00000308091.4 ENST00000453034.1 ENST00000420371.1 |
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr12_-_13105081 | 0.32 |
ENST00000541128.1
|
GPRC5D
|
G protein-coupled receptor, family C, group 5, member D |
| chr21_+_37529055 | 0.31 |
ENST00000270190.4
|
DOPEY2
|
dopey family member 2 |
| chr10_-_123687497 | 0.31 |
ENST00000369040.3
ENST00000224652.6 ENST00000369043.3 |
ATE1
|
arginyltransferase 1 |
| chr2_-_160568918 | 0.31 |
ENST00000453016.1
ENST00000607836.1 |
AC009961.3
|
AC009961.3 |
| chr10_-_103347883 | 0.31 |
ENST00000339310.3
ENST00000370158.3 ENST00000299206.4 ENST00000456836.2 ENST00000413344.1 ENST00000429502.1 ENST00000430045.1 ENST00000370172.1 ENST00000436284.2 ENST00000370162.3 |
POLL
|
polymerase (DNA directed), lambda |
| chr20_-_1373606 | 0.31 |
ENST00000381715.1
ENST00000439640.2 ENST00000381719.3 |
FKBP1A
|
FK506 binding protein 1A, 12kDa |
| chr10_+_35416223 | 0.31 |
ENST00000489321.1
ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM
|
cAMP responsive element modulator |
| chr1_-_21113105 | 0.30 |
ENST00000375000.1
ENST00000419490.1 ENST00000414993.1 ENST00000443615.1 ENST00000312239.5 |
HP1BP3
|
heterochromatin protein 1, binding protein 3 |
| chr6_-_79787902 | 0.30 |
ENST00000275034.4
|
PHIP
|
pleckstrin homology domain interacting protein |
| chr15_-_50411412 | 0.30 |
ENST00000284509.6
|
ATP8B4
|
ATPase, class I, type 8B, member 4 |
| chr11_-_8615488 | 0.30 |
ENST00000315204.1
ENST00000396672.1 ENST00000396673.1 |
STK33
|
serine/threonine kinase 33 |
| chr6_-_31550192 | 0.30 |
ENST00000429299.2
ENST00000446745.2 |
LTB
|
lymphotoxin beta (TNF superfamily, member 3) |
| chr5_-_133968459 | 0.29 |
ENST00000505758.1
ENST00000439578.1 ENST00000502286.1 |
SAR1B
|
SAR1 homolog B (S. cerevisiae) |
| chr19_+_55999916 | 0.28 |
ENST00000587166.1
ENST00000389623.6 |
SSC5D
|
scavenger receptor cysteine rich domain containing (5 domains) |
| chr7_-_84121858 | 0.28 |
ENST00000448879.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr20_-_62710832 | 0.28 |
ENST00000395042.1
|
RGS19
|
regulator of G-protein signaling 19 |
| chr1_+_95286151 | 0.28 |
ENST00000467909.1
ENST00000422520.2 ENST00000532427.1 |
SLC44A3
|
solute carrier family 44, member 3 |
| chr2_+_223916862 | 0.28 |
ENST00000604125.1
|
KCNE4
|
potassium voltage-gated channel, Isk-related family, member 4 |
| chr20_+_34802295 | 0.28 |
ENST00000432603.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr16_+_31404624 | 0.28 |
ENST00000389202.2
|
ITGAD
|
integrin, alpha D |
| chr14_+_105927191 | 0.28 |
ENST00000550551.1
|
MTA1
|
metastasis associated 1 |
| chr14_-_92414055 | 0.28 |
ENST00000342058.4
|
FBLN5
|
fibulin 5 |
| chr2_-_239197201 | 0.27 |
ENST00000254658.3
|
PER2
|
period circadian clock 2 |
| chr8_-_91657740 | 0.27 |
ENST00000422900.1
|
TMEM64
|
transmembrane protein 64 |
| chr9_-_88896977 | 0.27 |
ENST00000311534.6
|
ISCA1
|
iron-sulfur cluster assembly 1 |
| chr7_-_11871815 | 0.27 |
ENST00000423059.4
|
THSD7A
|
thrombospondin, type I, domain containing 7A |
| chr3_+_148847371 | 0.26 |
ENST00000296051.2
ENST00000460120.1 |
HPS3
|
Hermansky-Pudlak syndrome 3 |
| chr19_+_13988061 | 0.26 |
ENST00000339133.5
ENST00000397555.2 |
NANOS3
|
nanos homolog 3 (Drosophila) |
| chr1_-_38157877 | 0.26 |
ENST00000477060.1
ENST00000491981.1 ENST00000488137.1 |
C1orf109
|
chromosome 1 open reading frame 109 |
| chr7_-_120497178 | 0.26 |
ENST00000441017.1
ENST00000424710.1 ENST00000433758.1 |
TSPAN12
|
tetraspanin 12 |
| chrX_+_10031499 | 0.26 |
ENST00000454666.1
|
WWC3
|
WWC family member 3 |
| chr1_-_220445757 | 0.26 |
ENST00000358951.2
|
RAB3GAP2
|
RAB3 GTPase activating protein subunit 2 (non-catalytic) |
| chr11_-_8615687 | 0.26 |
ENST00000534493.1
ENST00000422559.2 |
STK33
|
serine/threonine kinase 33 |
| chr16_+_28996416 | 0.25 |
ENST00000395456.2
ENST00000454369.2 |
LAT
|
linker for activation of T cells |
| chr20_-_1373682 | 0.25 |
ENST00000381724.3
|
FKBP1A
|
FK506 binding protein 1A, 12kDa |
| chrX_+_38420623 | 0.25 |
ENST00000378482.2
|
TSPAN7
|
tetraspanin 7 |
| chr12_-_121972556 | 0.25 |
ENST00000545022.1
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr1_-_108231101 | 0.25 |
ENST00000544443.1
ENST00000415432.2 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr1_-_92764523 | 0.25 |
ENST00000370360.3
ENST00000534881.1 |
GLMN
|
glomulin, FKBP associated protein |
| chr11_+_18344106 | 0.25 |
ENST00000534641.1
ENST00000525831.1 ENST00000265963.4 |
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa |
| chr18_+_29077990 | 0.24 |
ENST00000261590.8
|
DSG2
|
desmoglein 2 |
| chr5_+_35852797 | 0.24 |
ENST00000508941.1
|
IL7R
|
interleukin 7 receptor |
| chr1_-_241803649 | 0.24 |
ENST00000366554.2
|
OPN3
|
opsin 3 |
| chr9_+_86595626 | 0.24 |
ENST00000445877.1
ENST00000325875.3 |
RMI1
|
RecQ mediated genome instability 1 |
| chr6_-_32920794 | 0.24 |
ENST00000395305.3
ENST00000395303.3 ENST00000374843.4 ENST00000429234.1 |
HLA-DMA
XXbac-BPG181M17.5
|
major histocompatibility complex, class II, DM alpha Uncharacterized protein |
| chr8_+_23145594 | 0.24 |
ENST00000519952.1
ENST00000518840.1 |
R3HCC1
|
R3H domain and coiled-coil containing 1 |
| chr19_-_1876156 | 0.24 |
ENST00000565797.1
|
CTB-31O20.2
|
CTB-31O20.2 |
| chr1_-_74663825 | 0.24 |
ENST00000370911.3
ENST00000370909.2 ENST00000354431.4 |
LRRIQ3
|
leucine-rich repeats and IQ motif containing 3 |
| chr10_-_123687943 | 0.24 |
ENST00000540606.1
ENST00000455628.1 |
ATE1
|
arginyltransferase 1 |
| chr3_+_151986709 | 0.24 |
ENST00000495875.2
ENST00000493459.1 ENST00000324210.5 ENST00000459747.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr14_+_35452169 | 0.23 |
ENST00000555557.1
|
SRP54
|
signal recognition particle 54kDa |
| chr7_-_99573640 | 0.23 |
ENST00000411734.1
|
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding |
| chr1_+_10509971 | 0.23 |
ENST00000320498.4
|
CORT
|
cortistatin |
| chr5_+_95998673 | 0.23 |
ENST00000514845.1
|
CAST
|
calpastatin |
| chr2_-_204400113 | 0.23 |
ENST00000319170.5
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr11_-_34938039 | 0.23 |
ENST00000395787.3
|
APIP
|
APAF1 interacting protein |
| chr10_-_124768300 | 0.23 |
ENST00000368886.5
|
IKZF5
|
IKAROS family zinc finger 5 (Pegasus) |
| chrX_+_38420783 | 0.23 |
ENST00000422612.2
ENST00000286824.6 ENST00000545599.1 |
TSPAN7
|
tetraspanin 7 |
| chr3_+_124355944 | 0.22 |
ENST00000459915.1
|
KALRN
|
kalirin, RhoGEF kinase |
| chr22_+_19706958 | 0.22 |
ENST00000395109.2
|
SEPT5
|
septin 5 |
| chr14_+_65170820 | 0.22 |
ENST00000555982.1
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr18_+_56531584 | 0.22 |
ENST00000590287.1
|
ZNF532
|
zinc finger protein 532 |
| chr11_+_93474757 | 0.22 |
ENST00000528288.1
|
C11orf54
|
chromosome 11 open reading frame 54 |
| chr1_+_93297622 | 0.22 |
ENST00000315741.5
|
RPL5
|
ribosomal protein L5 |
| chr19_+_840963 | 0.22 |
ENST00000234347.5
|
PRTN3
|
proteinase 3 |
| chr14_+_35452104 | 0.21 |
ENST00000216774.6
ENST00000546080.1 |
SRP54
|
signal recognition particle 54kDa |
| chr8_+_123875624 | 0.21 |
ENST00000534247.1
|
ZHX2
|
zinc fingers and homeoboxes 2 |
| chr4_+_140222609 | 0.21 |
ENST00000296543.5
ENST00000398947.1 |
NAA15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr1_+_236958554 | 0.21 |
ENST00000366577.5
ENST00000418145.2 |
MTR
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr14_+_50779071 | 0.21 |
ENST00000426751.2
|
ATP5S
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit s (factor B) |
| chr11_+_93474786 | 0.21 |
ENST00000331239.4
ENST00000533585.1 ENST00000528099.1 ENST00000354421.3 ENST00000540113.1 ENST00000530620.1 ENST00000527003.1 ENST00000531650.1 ENST00000530279.1 |
C11orf54
|
chromosome 11 open reading frame 54 |
| chr4_+_48485341 | 0.21 |
ENST00000273861.4
|
SLC10A4
|
solute carrier family 10, member 4 |
| chr12_-_89919220 | 0.21 |
ENST00000549035.1
ENST00000393179.4 |
POC1B
|
POC1 centriolar protein B |
| chr10_+_99205959 | 0.20 |
ENST00000352634.4
ENST00000353979.3 ENST00000370842.2 ENST00000345745.5 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chr11_-_93474645 | 0.20 |
ENST00000532455.1
|
TAF1D
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, D, 41kDa |
| chr11_-_8615720 | 0.20 |
ENST00000358872.3
ENST00000454443.2 |
STK33
|
serine/threonine kinase 33 |
| chr1_+_24645865 | 0.20 |
ENST00000342072.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr9_+_100395891 | 0.20 |
ENST00000375147.3
|
NCBP1
|
nuclear cap binding protein subunit 1, 80kDa |
| chr12_-_122107549 | 0.20 |
ENST00000355329.3
|
MORN3
|
MORN repeat containing 3 |
| chr4_-_75719896 | 0.20 |
ENST00000395743.3
|
BTC
|
betacellulin |
| chr2_+_102927962 | 0.20 |
ENST00000233954.1
ENST00000393393.3 ENST00000410040.1 |
IL1RL1
IL18R1
|
interleukin 1 receptor-like 1 interleukin 18 receptor 1 |
| chr14_+_54863739 | 0.20 |
ENST00000541304.1
|
CDKN3
|
cyclin-dependent kinase inhibitor 3 |
| chr19_-_51920873 | 0.20 |
ENST00000441969.3
ENST00000525998.1 ENST00000436984.2 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chr21_-_16437255 | 0.20 |
ENST00000400199.1
ENST00000400202.1 |
NRIP1
|
nuclear receptor interacting protein 1 |
| chr17_-_70053866 | 0.20 |
ENST00000540802.1
|
RP11-84E24.2
|
RP11-84E24.2 |
| chr11_+_18343800 | 0.19 |
ENST00000453096.2
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa |
| chr21_-_16437126 | 0.19 |
ENST00000318948.4
|
NRIP1
|
nuclear receptor interacting protein 1 |
| chr1_+_24646002 | 0.19 |
ENST00000356046.2
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr8_+_18067602 | 0.19 |
ENST00000307719.4
ENST00000545197.1 ENST00000539092.1 ENST00000541942.1 ENST00000518029.1 |
NAT1
|
N-acetyltransferase 1 (arylamine N-acetyltransferase) |
| chr12_-_90024360 | 0.19 |
ENST00000393164.2
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr14_-_54955721 | 0.19 |
ENST00000554908.1
|
GMFB
|
glia maturation factor, beta |
| chr14_+_54863682 | 0.19 |
ENST00000543789.2
ENST00000442975.2 ENST00000458126.2 ENST00000556102.2 |
CDKN3
|
cyclin-dependent kinase inhibitor 3 |
| chr16_-_16317321 | 0.19 |
ENST00000205557.7
ENST00000575728.1 |
ABCC6
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6 |
| chr20_+_56884752 | 0.19 |
ENST00000244040.3
|
RAB22A
|
RAB22A, member RAS oncogene family |
| chr2_-_160568529 | 0.19 |
ENST00000418770.1
|
AC009961.3
|
AC009961.3 |
| chr20_-_13765526 | 0.19 |
ENST00000202816.1
|
ESF1
|
ESF1, nucleolar pre-rRNA processing protein, homolog (S. cerevisiae) |
| chr4_-_147442817 | 0.19 |
ENST00000507030.1
|
SLC10A7
|
solute carrier family 10, member 7 |
| chr3_-_194373831 | 0.19 |
ENST00000437613.1
|
LSG1
|
large 60S subunit nuclear export GTPase 1 |
| chr15_+_67835517 | 0.19 |
ENST00000395476.2
|
MAP2K5
|
mitogen-activated protein kinase kinase 5 |
| chr1_+_24645807 | 0.19 |
ENST00000361548.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr11_-_102323740 | 0.19 |
ENST00000398136.2
|
TMEM123
|
transmembrane protein 123 |
| chr1_-_26324534 | 0.19 |
ENST00000374284.1
ENST00000441420.1 ENST00000374282.3 |
PAFAH2
|
platelet-activating factor acetylhydrolase 2, 40kDa |
| chr15_+_42694573 | 0.19 |
ENST00000397200.4
ENST00000569827.1 |
CAPN3
|
calpain 3, (p94) |
| chr11_+_111808119 | 0.18 |
ENST00000531396.1
|
DIXDC1
|
DIX domain containing 1 |
| chr8_-_55014415 | 0.18 |
ENST00000522007.1
ENST00000521898.1 ENST00000518546.1 ENST00000316963.3 |
LYPLA1
|
lysophospholipase I |
| chr2_-_175869936 | 0.18 |
ENST00000409900.3
|
CHN1
|
chimerin 1 |
| chr9_+_116225999 | 0.18 |
ENST00000317613.6
|
RGS3
|
regulator of G-protein signaling 3 |
| chr7_+_139528952 | 0.18 |
ENST00000416849.2
ENST00000436047.2 ENST00000414508.2 ENST00000448866.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr6_+_13272904 | 0.18 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr16_-_28223166 | 0.18 |
ENST00000304658.5
|
XPO6
|
exportin 6 |
| chr9_+_117085336 | 0.18 |
ENST00000259396.8
ENST00000538816.1 |
ORM1
|
orosomucoid 1 |
| chr5_+_68710906 | 0.18 |
ENST00000325631.5
ENST00000454295.2 |
MARVELD2
|
MARVEL domain containing 2 |
| chr4_+_165675269 | 0.18 |
ENST00000507311.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr19_-_11266471 | 0.18 |
ENST00000592540.1
|
SPC24
|
SPC24, NDC80 kinetochore complex component |
| chr5_-_133968529 | 0.18 |
ENST00000402673.2
|
SAR1B
|
SAR1 homolog B (S. cerevisiae) |
| chr7_+_139529040 | 0.18 |
ENST00000455353.1
ENST00000458722.1 ENST00000411653.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr12_+_85673868 | 0.18 |
ENST00000316824.3
|
ALX1
|
ALX homeobox 1 |
| chr10_-_79686284 | 0.18 |
ENST00000372391.2
ENST00000372388.2 |
DLG5
|
discs, large homolog 5 (Drosophila) |
| chr1_-_151762900 | 0.18 |
ENST00000440583.2
|
TDRKH
|
tudor and KH domain containing |
| chr19_-_44079611 | 0.18 |
ENST00000594107.1
ENST00000543982.1 |
XRCC1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr10_-_60027642 | 0.18 |
ENST00000373935.3
|
IPMK
|
inositol polyphosphate multikinase |
| chr14_+_70234642 | 0.17 |
ENST00000555349.1
ENST00000554021.1 |
SRSF5
|
serine/arginine-rich splicing factor 5 |
| chr2_-_190044480 | 0.17 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr5_-_137610254 | 0.17 |
ENST00000378362.3
|
GFRA3
|
GDNF family receptor alpha 3 |
| chr3_-_179322416 | 0.17 |
ENST00000259038.2
|
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr3_+_132036207 | 0.17 |
ENST00000336375.5
ENST00000495911.1 |
ACPP
|
acid phosphatase, prostate |
| chr3_+_108308513 | 0.17 |
ENST00000361582.3
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr15_+_75080883 | 0.17 |
ENST00000567571.1
|
CSK
|
c-src tyrosine kinase |
| chr7_-_32111009 | 0.17 |
ENST00000396184.3
ENST00000396189.2 ENST00000321453.7 |
PDE1C
|
phosphodiesterase 1C, calmodulin-dependent 70kDa |
| chr11_-_1036706 | 0.17 |
ENST00000421673.2
|
MUC6
|
mucin 6, oligomeric mucus/gel-forming |
| chr19_-_46405861 | 0.17 |
ENST00000322217.5
|
MYPOP
|
Myb-related transcription factor, partner of profilin |
| chr8_+_57124245 | 0.17 |
ENST00000521831.1
ENST00000355315.3 ENST00000303759.3 ENST00000517636.1 ENST00000517933.1 ENST00000518801.1 ENST00000523975.1 ENST00000396723.5 ENST00000523061.1 ENST00000521524.1 |
CHCHD7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr10_+_99205894 | 0.17 |
ENST00000370854.3
ENST00000393760.1 ENST00000414567.1 ENST00000370846.4 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chr7_+_99971129 | 0.17 |
ENST00000394000.2
ENST00000350573.2 |
PILRA
|
paired immunoglobin-like type 2 receptor alpha |
| chr1_+_155829341 | 0.16 |
ENST00000539162.1
|
SYT11
|
synaptotagmin XI |
| chr7_+_12544025 | 0.16 |
ENST00000443874.1
ENST00000424453.1 |
AC005281.1
|
AC005281.1 |
| chr2_+_63816295 | 0.16 |
ENST00000539945.1
ENST00000544381.1 |
MDH1
|
malate dehydrogenase 1, NAD (soluble) |
| chr15_+_91478493 | 0.16 |
ENST00000418476.2
|
UNC45A
|
unc-45 homolog A (C. elegans) |
Network of associatons between targets according to the STRING database.
First level regulatory network of TLX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.2 | 0.7 | GO:0070839 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) divalent metal ion export(GO:0070839) |
| 0.1 | 0.6 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.1 | 0.4 | GO:0002514 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.1 | 0.4 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.1 | 0.4 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.1 | 0.5 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.1 | 0.7 | GO:0019427 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.1 | 0.4 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.1 | 0.5 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.1 | 0.7 | GO:0021546 | rhombomere development(GO:0021546) |
| 0.1 | 0.3 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.1 | 0.3 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 0.4 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 0.6 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.4 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.1 | 0.8 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.1 | 0.3 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.1 | 0.6 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.2 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.1 | 0.2 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.1 | 0.4 | GO:1901569 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.1 | 0.3 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.1 | 0.3 | GO:1990927 | negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.1 | 0.5 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.3 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.1 | 0.2 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.1 | 0.7 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.1 | 0.2 | GO:1904875 | regulation of DNA ligase activity(GO:1904875) |
| 0.1 | 0.8 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.3 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.2 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.0 | 0.3 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.3 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.0 | 0.5 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.3 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.2 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.0 | 0.4 | GO:0070269 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) pyroptosis(GO:0070269) |
| 0.0 | 0.4 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.6 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.7 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.4 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.2 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.1 | GO:0080121 | coenzyme A transport(GO:0015880) coenzyme A transmembrane transport(GO:0035349) adenosine 3',5'-bisphosphate transmembrane transport(GO:0071106) AMP transport(GO:0080121) |
| 0.0 | 1.1 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.8 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.8 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.2 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.1 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.0 | 0.1 | GO:2000584 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.6 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.3 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.2 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.1 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.3 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.2 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.3 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.1 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.2 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.0 | 0.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:1905216 | positive regulation of RNA binding(GO:1905216) |
| 0.0 | 0.3 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.2 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.1 | GO:1903980 | positive regulation of microglial cell activation(GO:1903980) |
| 0.0 | 0.1 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.0 | 0.4 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.1 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.0 | 0.2 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.2 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.0 | 0.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.3 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.4 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.4 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.4 | GO:0046174 | polyol catabolic process(GO:0046174) |
| 0.0 | 0.1 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.2 | GO:0071351 | interleukin-18-mediated signaling pathway(GO:0035655) cellular response to interleukin-18(GO:0071351) |
| 0.0 | 0.3 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.0 | 0.2 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 0.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.1 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.1 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.0 | GO:0071503 | response to heparin(GO:0071503) |
| 0.0 | 0.0 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.0 | 0.0 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.3 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.0 | 0.2 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.3 | GO:0098743 | cell aggregation(GO:0098743) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.0 | 0.2 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.1 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.9 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.4 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.1 | 0.2 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 0.2 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.1 | 0.3 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.3 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.1 | 0.2 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 0.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.3 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.4 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.4 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.4 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.4 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.1 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.4 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.3 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.4 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.0 | 0.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.1 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.4 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0004057 | arginyltransferase activity(GO:0004057) |
| 0.1 | 0.4 | GO:0052870 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.1 | 0.3 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 0.8 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.4 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 0.4 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.1 | 0.4 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.1 | 0.7 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 0.3 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.1 | 0.2 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.1 | 0.2 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.1 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.7 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.2 | GO:0047860 | diiodophenylpyruvate reductase activity(GO:0047860) |
| 0.0 | 0.4 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.2 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0003955 | NAD(P)H dehydrogenase (quinone) activity(GO:0003955) |
| 0.0 | 0.2 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.0 | 0.3 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.0 | 0.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.8 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.3 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.3 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.1 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.3 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.1 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.3 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.4 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.4 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.5 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.4 | GO:0055103 | ligase regulator activity(GO:0055103) |
| 0.0 | 0.1 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.0 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.1 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.7 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.3 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0070325 | lipoprotein particle receptor binding(GO:0070325) |
| 0.0 | 0.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.5 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 1.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.3 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 1.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.7 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.6 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.4 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |