Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
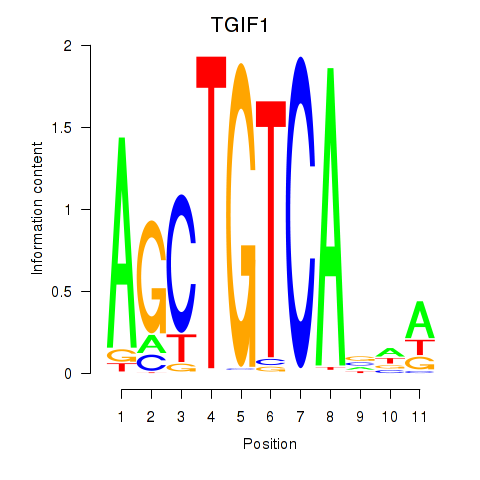
Results for TGIF1
Z-value: 0.25
Transcription factors associated with TGIF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TGIF1
|
ENSG00000177426.16 | TGFB induced factor homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TGIF1 | hg19_v2_chr18_+_3451584_3451606 | -0.97 | 2.8e-02 | Click! |
Activity profile of TGIF1 motif
Sorted Z-values of TGIF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_11907829 | 0.27 |
ENST00000376480.3
|
NPPA
|
natriuretic peptide A |
| chr10_-_10504285 | 0.23 |
ENST00000602311.1
|
RP11-271F18.4
|
RP11-271F18.4 |
| chr15_+_75335604 | 0.15 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr3_-_127541194 | 0.14 |
ENST00000453507.2
|
MGLL
|
monoglyceride lipase |
| chr6_+_131521120 | 0.14 |
ENST00000537868.1
|
AKAP7
|
A kinase (PRKA) anchor protein 7 |
| chr20_-_31124186 | 0.14 |
ENST00000375678.3
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr19_-_49567124 | 0.13 |
ENST00000301411.3
|
NTF4
|
neurotrophin 4 |
| chr12_+_25348186 | 0.13 |
ENST00000555711.1
ENST00000556885.1 ENST00000554266.1 ENST00000556351.1 ENST00000556927.1 ENST00000556402.1 ENST00000553788.1 |
LYRM5
|
LYR motif containing 5 |
| chr22_-_36013368 | 0.11 |
ENST00000442617.1
ENST00000397326.2 ENST00000397328.1 ENST00000451685.1 |
MB
|
myoglobin |
| chr13_-_34250861 | 0.11 |
ENST00000445227.1
ENST00000454681.2 |
RP11-141M1.3
|
RP11-141M1.3 |
| chr7_-_50628745 | 0.10 |
ENST00000380984.4
ENST00000357936.5 ENST00000426377.1 |
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase) |
| chr9_-_33447584 | 0.10 |
ENST00000297991.4
|
AQP3
|
aquaporin 3 (Gill blood group) |
| chr17_+_57233087 | 0.10 |
ENST00000578777.1
ENST00000577457.1 ENST00000582995.1 |
PRR11
|
proline rich 11 |
| chr6_-_79787902 | 0.10 |
ENST00000275034.4
|
PHIP
|
pleckstrin homology domain interacting protein |
| chr4_-_105416039 | 0.10 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr3_-_123168551 | 0.09 |
ENST00000462833.1
|
ADCY5
|
adenylate cyclase 5 |
| chr17_-_28257080 | 0.09 |
ENST00000579954.1
ENST00000540801.1 ENST00000269033.3 ENST00000590153.1 ENST00000582084.1 |
SSH2
|
slingshot protein phosphatase 2 |
| chr7_+_66800928 | 0.09 |
ENST00000430244.1
|
RP11-166O4.5
|
RP11-166O4.5 |
| chr4_+_119200215 | 0.09 |
ENST00000602573.1
|
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr3_+_107318157 | 0.09 |
ENST00000406780.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chr3_+_38017264 | 0.09 |
ENST00000436654.1
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr16_+_66600294 | 0.09 |
ENST00000535705.1
ENST00000332695.7 ENST00000336328.6 ENST00000528324.1 ENST00000531885.1 ENST00000529506.1 ENST00000457188.2 ENST00000533666.1 ENST00000533953.1 ENST00000379500.2 ENST00000328020.6 |
CMTM1
|
CKLF-like MARVEL transmembrane domain containing 1 |
| chr2_-_3595547 | 0.09 |
ENST00000438485.1
|
RP13-512J5.1
|
Uncharacterized protein |
| chr16_+_2198604 | 0.08 |
ENST00000210187.6
|
RAB26
|
RAB26, member RAS oncogene family |
| chr1_-_150979333 | 0.08 |
ENST00000312210.5
|
FAM63A
|
family with sequence similarity 63, member A |
| chr8_+_71485681 | 0.08 |
ENST00000391684.1
|
AC120194.1
|
AC120194.1 |
| chr11_+_63870660 | 0.08 |
ENST00000246841.3
|
FLRT1
|
fibronectin leucine rich transmembrane protein 1 |
| chr5_-_173043591 | 0.08 |
ENST00000285908.5
ENST00000480951.1 ENST00000311086.4 |
BOD1
|
biorientation of chromosomes in cell division 1 |
| chr17_+_74734052 | 0.08 |
ENST00000590514.1
|
MFSD11
|
major facilitator superfamily domain containing 11 |
| chr2_+_27719697 | 0.08 |
ENST00000264717.2
ENST00000424318.2 |
GCKR
|
glucokinase (hexokinase 4) regulator |
| chr4_+_14113592 | 0.08 |
ENST00000502759.1
ENST00000511200.1 ENST00000512754.1 ENST00000506739.1 |
LINC01085
|
long intergenic non-protein coding RNA 1085 |
| chr10_+_94608245 | 0.08 |
ENST00000443748.2
ENST00000260762.6 |
EXOC6
|
exocyst complex component 6 |
| chr1_-_241799232 | 0.08 |
ENST00000366553.1
|
CHML
|
choroideremia-like (Rab escort protein 2) |
| chr17_+_57232690 | 0.08 |
ENST00000262293.4
|
PRR11
|
proline rich 11 |
| chr10_-_90751038 | 0.08 |
ENST00000458159.1
ENST00000415557.1 ENST00000458208.1 |
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr6_+_80714318 | 0.08 |
ENST00000369798.2
|
TTK
|
TTK protein kinase |
| chr17_+_4618734 | 0.08 |
ENST00000571206.1
|
ARRB2
|
arrestin, beta 2 |
| chr20_+_4667094 | 0.08 |
ENST00000424424.1
ENST00000457586.1 |
PRNP
|
prion protein |
| chr13_+_50570019 | 0.07 |
ENST00000442421.1
|
TRIM13
|
tripartite motif containing 13 |
| chr22_+_37959647 | 0.07 |
ENST00000415670.1
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chr17_-_39156138 | 0.07 |
ENST00000391587.1
|
KRTAP3-2
|
keratin associated protein 3-2 |
| chr12_+_19593515 | 0.07 |
ENST00000360995.4
|
AEBP2
|
AE binding protein 2 |
| chr5_-_139930713 | 0.07 |
ENST00000602657.1
|
SRA1
|
steroid receptor RNA activator 1 |
| chr17_+_76210367 | 0.07 |
ENST00000592734.1
ENST00000587746.1 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chr3_-_49066811 | 0.07 |
ENST00000442157.1
ENST00000326739.4 |
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr2_+_33359687 | 0.07 |
ENST00000402934.1
ENST00000404525.1 ENST00000407925.1 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr3_-_142166846 | 0.07 |
ENST00000463916.1
ENST00000544157.1 |
XRN1
|
5'-3' exoribonuclease 1 |
| chr9_-_36276966 | 0.07 |
ENST00000543356.2
ENST00000396594.3 |
GNE
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr2_+_27505260 | 0.07 |
ENST00000380075.2
ENST00000296098.4 |
TRIM54
|
tripartite motif containing 54 |
| chr2_-_74779744 | 0.07 |
ENST00000409249.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr12_-_62653903 | 0.07 |
ENST00000552075.1
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chr12_+_25348139 | 0.07 |
ENST00000557540.2
ENST00000381356.4 |
LYRM5
|
LYR motif containing 5 |
| chr7_+_76101379 | 0.07 |
ENST00000429179.1
|
DTX2
|
deltex homolog 2 (Drosophila) |
| chr12_+_65672702 | 0.07 |
ENST00000538045.1
ENST00000535239.1 |
MSRB3
|
methionine sulfoxide reductase B3 |
| chr9_-_130635741 | 0.06 |
ENST00000223836.10
|
AK1
|
adenylate kinase 1 |
| chr12_-_57443886 | 0.06 |
ENST00000300119.3
|
MYO1A
|
myosin IA |
| chr1_-_193075180 | 0.06 |
ENST00000367440.3
|
GLRX2
|
glutaredoxin 2 |
| chr3_-_127541679 | 0.06 |
ENST00000265052.5
|
MGLL
|
monoglyceride lipase |
| chr5_-_66942617 | 0.06 |
ENST00000507298.1
|
RP11-83M16.5
|
RP11-83M16.5 |
| chr17_-_57232525 | 0.06 |
ENST00000583380.1
ENST00000580541.1 ENST00000578105.1 ENST00000437036.2 |
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr1_+_18807424 | 0.06 |
ENST00000400664.1
|
KLHDC7A
|
kelch domain containing 7A |
| chr9_-_35812140 | 0.06 |
ENST00000497810.1
ENST00000396638.2 ENST00000484764.1 |
SPAG8
|
sperm associated antigen 8 |
| chr3_+_188817891 | 0.06 |
ENST00000412373.1
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr12_-_71512027 | 0.06 |
ENST00000549357.1
|
CTD-2021H9.3
|
Uncharacterized protein |
| chr12_-_39836772 | 0.06 |
ENST00000541463.2
ENST00000361418.5 ENST00000544797.2 |
KIF21A
|
kinesin family member 21A |
| chr4_-_114682936 | 0.06 |
ENST00000454265.2
ENST00000429180.1 ENST00000418639.2 ENST00000394526.2 ENST00000296402.5 |
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta |
| chr18_-_55253989 | 0.06 |
ENST00000262093.5
|
FECH
|
ferrochelatase |
| chr2_+_33359646 | 0.06 |
ENST00000390003.4
ENST00000418533.2 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr4_-_16228083 | 0.06 |
ENST00000399920.3
|
TAPT1
|
transmembrane anterior posterior transformation 1 |
| chr19_-_10946871 | 0.06 |
ENST00000589638.1
|
TMED1
|
transmembrane emp24 protein transport domain containing 1 |
| chr2_-_152589670 | 0.06 |
ENST00000604864.1
ENST00000603639.1 |
NEB
|
nebulin |
| chr3_-_129513259 | 0.06 |
ENST00000329333.5
|
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr6_-_72130472 | 0.06 |
ENST00000426635.2
|
LINC00472
|
long intergenic non-protein coding RNA 472 |
| chr1_-_150978953 | 0.06 |
ENST00000493834.2
|
FAM63A
|
family with sequence similarity 63, member A |
| chr3_-_127542021 | 0.06 |
ENST00000434178.2
|
MGLL
|
monoglyceride lipase |
| chr6_+_24126350 | 0.05 |
ENST00000378491.4
ENST00000378478.1 ENST00000378477.2 |
NRSN1
|
neurensin 1 |
| chr21_+_30502806 | 0.05 |
ENST00000399928.1
ENST00000399926.1 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr9_+_34652164 | 0.05 |
ENST00000441545.2
ENST00000553620.1 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr3_-_127542051 | 0.05 |
ENST00000398104.1
|
MGLL
|
monoglyceride lipase |
| chr1_-_155959280 | 0.05 |
ENST00000495070.2
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr22_-_37640277 | 0.05 |
ENST00000401529.3
ENST00000249071.6 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr19_-_10946949 | 0.05 |
ENST00000214869.2
ENST00000591695.1 |
TMED1
|
transmembrane emp24 protein transport domain containing 1 |
| chr13_-_48575376 | 0.05 |
ENST00000434484.1
|
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr16_-_3030407 | 0.05 |
ENST00000431515.2
ENST00000574385.1 ENST00000576268.1 ENST00000574730.1 ENST00000575632.1 ENST00000573944.1 ENST00000262300.8 |
PKMYT1
|
protein kinase, membrane associated tyrosine/threonine 1 |
| chr14_-_23299009 | 0.05 |
ENST00000488800.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr22_+_50946645 | 0.05 |
ENST00000420993.2
ENST00000395698.3 ENST00000395701.3 ENST00000523045.1 ENST00000299821.11 |
NCAPH2
|
non-SMC condensin II complex, subunit H2 |
| chr11_-_6426635 | 0.05 |
ENST00000608645.1
ENST00000608394.1 ENST00000529519.1 |
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr8_-_41522779 | 0.05 |
ENST00000522231.1
ENST00000314214.8 ENST00000348036.4 ENST00000457297.1 ENST00000522543.1 |
ANK1
|
ankyrin 1, erythrocytic |
| chr15_+_36887069 | 0.05 |
ENST00000566807.1
ENST00000567389.1 ENST00000562877.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr2_+_201936707 | 0.05 |
ENST00000433898.1
ENST00000454214.1 |
NDUFB3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr10_+_60028818 | 0.05 |
ENST00000333926.5
|
CISD1
|
CDGSH iron sulfur domain 1 |
| chr15_-_37392086 | 0.05 |
ENST00000561208.1
|
MEIS2
|
Meis homeobox 2 |
| chr19_+_19030497 | 0.05 |
ENST00000438170.2
|
DDX49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr17_-_56296580 | 0.05 |
ENST00000313863.6
ENST00000546108.1 ENST00000337050.7 ENST00000393119.2 |
MKS1
|
Meckel syndrome, type 1 |
| chr13_-_96329048 | 0.05 |
ENST00000606011.1
ENST00000499499.2 |
DNAJC3-AS1
|
DNAJC3 antisense RNA 1 (head to head) |
| chr9_+_125512019 | 0.05 |
ENST00000373684.1
ENST00000304720.2 |
OR1L6
|
olfactory receptor, family 1, subfamily L, member 6 |
| chr11_+_6411670 | 0.05 |
ENST00000530395.1
ENST00000527275.1 |
SMPD1
|
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr2_+_128177458 | 0.05 |
ENST00000409048.1
ENST00000422777.3 |
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr5_+_139055055 | 0.05 |
ENST00000511457.1
|
CXXC5
|
CXXC finger protein 5 |
| chr17_-_27418537 | 0.05 |
ENST00000408971.2
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr8_+_99076750 | 0.05 |
ENST00000545282.1
|
C8orf47
|
chromosome 8 open reading frame 47 |
| chr14_+_70078303 | 0.05 |
ENST00000342745.4
|
KIAA0247
|
KIAA0247 |
| chr12_-_76461249 | 0.05 |
ENST00000551524.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr15_+_93426514 | 0.05 |
ENST00000556722.1
|
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr12_+_49621658 | 0.05 |
ENST00000541364.1
|
TUBA1C
|
tubulin, alpha 1c |
| chr15_+_40733387 | 0.05 |
ENST00000416165.1
|
BAHD1
|
bromo adjacent homology domain containing 1 |
| chr21_+_44394742 | 0.05 |
ENST00000432907.2
|
PKNOX1
|
PBX/knotted 1 homeobox 1 |
| chr18_-_55253900 | 0.05 |
ENST00000585747.1
ENST00000592699.1 |
FECH
|
ferrochelatase |
| chr1_-_65533390 | 0.05 |
ENST00000448344.1
|
RP4-535B20.1
|
RP4-535B20.1 |
| chr3_-_52719810 | 0.05 |
ENST00000424867.1
ENST00000394830.3 ENST00000431678.1 ENST00000450271.1 |
PBRM1
|
polybromo 1 |
| chr12_-_39837192 | 0.05 |
ENST00000361961.3
ENST00000395670.3 |
KIF21A
|
kinesin family member 21A |
| chr1_-_146696901 | 0.05 |
ENST00000369272.3
ENST00000441068.2 |
FMO5
|
flavin containing monooxygenase 5 |
| chr5_+_180257951 | 0.04 |
ENST00000501855.2
|
LINC00847
|
long intergenic non-protein coding RNA 847 |
| chr20_+_4666882 | 0.04 |
ENST00000379440.4
ENST00000430350.2 |
PRNP
|
prion protein |
| chr20_+_34802295 | 0.04 |
ENST00000432603.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr18_+_32556892 | 0.04 |
ENST00000591734.1
ENST00000413393.1 ENST00000589180.1 ENST00000587359.1 |
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr19_-_47164386 | 0.04 |
ENST00000391916.2
ENST00000410105.2 |
DACT3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr14_+_63671105 | 0.04 |
ENST00000316754.3
|
RHOJ
|
ras homolog family member J |
| chr12_+_104458235 | 0.04 |
ENST00000229330.4
|
HCFC2
|
host cell factor C2 |
| chr9_-_215744 | 0.04 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chr9_-_35812236 | 0.04 |
ENST00000340291.2
|
SPAG8
|
sperm associated antigen 8 |
| chr6_+_80714332 | 0.04 |
ENST00000502580.1
ENST00000511260.1 |
TTK
|
TTK protein kinase |
| chrX_+_47696337 | 0.04 |
ENST00000334937.4
ENST00000376950.4 |
ZNF81
|
zinc finger protein 81 |
| chr12_-_28122980 | 0.04 |
ENST00000395868.3
ENST00000534890.1 |
PTHLH
|
parathyroid hormone-like hormone |
| chr22_-_37640456 | 0.04 |
ENST00000405484.1
ENST00000441619.1 ENST00000406508.1 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr1_+_228870824 | 0.04 |
ENST00000366691.3
|
RHOU
|
ras homolog family member U |
| chr12_-_25348007 | 0.04 |
ENST00000354189.5
ENST00000545133.1 ENST00000554347.1 ENST00000395987.3 ENST00000320267.9 ENST00000395990.2 ENST00000537577.1 |
CASC1
|
cancer susceptibility candidate 1 |
| chr4_-_100871506 | 0.04 |
ENST00000296417.5
|
H2AFZ
|
H2A histone family, member Z |
| chr5_+_139055021 | 0.04 |
ENST00000502716.1
ENST00000503511.1 |
CXXC5
|
CXXC finger protein 5 |
| chr9_+_125027127 | 0.04 |
ENST00000441707.1
ENST00000373723.5 ENST00000373729.1 |
MRRF
|
mitochondrial ribosome recycling factor |
| chr1_-_53163992 | 0.04 |
ENST00000371538.3
|
SELRC1
|
cytochrome c oxidase assembly factor 7 |
| chr7_+_134551583 | 0.04 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr8_-_29120580 | 0.04 |
ENST00000524189.1
|
KIF13B
|
kinesin family member 13B |
| chr2_-_73460334 | 0.04 |
ENST00000258083.2
|
PRADC1
|
protease-associated domain containing 1 |
| chr4_-_175443943 | 0.04 |
ENST00000296522.6
|
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr9_+_131873659 | 0.04 |
ENST00000452489.2
ENST00000347048.4 ENST00000357197.4 ENST00000445241.1 ENST00000355007.3 ENST00000414331.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr12_+_4647950 | 0.04 |
ENST00000321524.7
ENST00000543041.1 ENST00000228843.9 ENST00000352618.4 ENST00000544927.1 |
RAD51AP1
|
RAD51 associated protein 1 |
| chr3_-_101232019 | 0.04 |
ENST00000394095.2
ENST00000394091.1 ENST00000394094.2 ENST00000358203.3 ENST00000348610.3 ENST00000314261.7 |
SENP7
|
SUMO1/sentrin specific peptidase 7 |
| chr10_+_94352956 | 0.04 |
ENST00000260731.3
|
KIF11
|
kinesin family member 11 |
| chr2_-_216003127 | 0.04 |
ENST00000412081.1
ENST00000272895.7 |
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr19_+_7953417 | 0.04 |
ENST00000600345.1
ENST00000598224.1 |
LRRC8E
|
leucine rich repeat containing 8 family, member E |
| chr2_+_239756671 | 0.04 |
ENST00000448943.2
|
TWIST2
|
twist family bHLH transcription factor 2 |
| chr4_-_114682597 | 0.04 |
ENST00000394524.3
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta |
| chr15_-_81195510 | 0.04 |
ENST00000561295.1
|
RP11-351M8.1
|
Uncharacterized protein |
| chr7_-_140155711 | 0.04 |
ENST00000463142.1
|
MKRN1
|
makorin ring finger protein 1 |
| chr4_-_110736505 | 0.04 |
ENST00000609440.1
|
RP11-602N24.3
|
RP11-602N24.3 |
| chr16_+_2880157 | 0.04 |
ENST00000382280.3
|
ZG16B
|
zymogen granule protein 16B |
| chr15_+_101389945 | 0.04 |
ENST00000561231.1
ENST00000559331.1 ENST00000558254.1 |
RP11-66B24.2
|
RP11-66B24.2 |
| chr11_-_2924720 | 0.04 |
ENST00000455942.2
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr1_-_91317072 | 0.04 |
ENST00000435649.2
ENST00000443802.1 |
RP4-665J23.1
|
RP4-665J23.1 |
| chr19_+_58111241 | 0.04 |
ENST00000597700.1
ENST00000332854.6 ENST00000597864.1 |
ZNF530
|
zinc finger protein 530 |
| chr12_-_49318715 | 0.04 |
ENST00000444214.2
|
FKBP11
|
FK506 binding protein 11, 19 kDa |
| chr22_-_40929812 | 0.03 |
ENST00000422851.1
|
MKL1
|
megakaryoblastic leukemia (translocation) 1 |
| chr12_-_88974236 | 0.03 |
ENST00000228280.5
ENST00000552044.1 ENST00000357116.4 |
KITLG
|
KIT ligand |
| chr11_-_119999539 | 0.03 |
ENST00000541857.1
|
TRIM29
|
tripartite motif containing 29 |
| chr16_+_27413483 | 0.03 |
ENST00000337929.3
ENST00000564089.1 |
IL21R
|
interleukin 21 receptor |
| chr9_+_131873591 | 0.03 |
ENST00000393370.2
ENST00000337738.1 ENST00000348141.5 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr6_+_37225540 | 0.03 |
ENST00000373491.3
|
TBC1D22B
|
TBC1 domain family, member 22B |
| chr3_-_27764190 | 0.03 |
ENST00000537516.1
|
EOMES
|
eomesodermin |
| chr18_+_3450161 | 0.03 |
ENST00000551402.1
ENST00000577543.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr5_+_139505520 | 0.03 |
ENST00000333305.3
|
IGIP
|
IgA-inducing protein |
| chrX_+_70443050 | 0.03 |
ENST00000361726.6
|
GJB1
|
gap junction protein, beta 1, 32kDa |
| chr15_-_82338460 | 0.03 |
ENST00000558133.1
ENST00000329713.4 |
MEX3B
|
mex-3 RNA binding family member B |
| chr9_+_134065506 | 0.03 |
ENST00000483497.2
|
NUP214
|
nucleoporin 214kDa |
| chr12_+_65672423 | 0.03 |
ENST00000355192.3
ENST00000308259.5 ENST00000540804.1 ENST00000535664.1 ENST00000541189.1 |
MSRB3
|
methionine sulfoxide reductase B3 |
| chr8_+_145438870 | 0.03 |
ENST00000527931.1
|
FAM203B
|
family with sequence similarity 203, member B |
| chr20_+_35807714 | 0.03 |
ENST00000373632.4
|
RPN2
|
ribophorin II |
| chr9_-_130637244 | 0.03 |
ENST00000373156.1
|
AK1
|
adenylate kinase 1 |
| chr1_-_204329013 | 0.03 |
ENST00000272203.3
ENST00000414478.1 |
PLEKHA6
|
pleckstrin homology domain containing, family A member 6 |
| chr9_-_134615443 | 0.03 |
ENST00000372195.1
|
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr5_+_180257974 | 0.03 |
ENST00000502162.2
ENST00000508309.1 ENST00000509921.1 |
LINC00847
|
long intergenic non-protein coding RNA 847 |
| chr2_+_223916862 | 0.03 |
ENST00000604125.1
|
KCNE4
|
potassium voltage-gated channel, Isk-related family, member 4 |
| chr22_-_39150947 | 0.03 |
ENST00000411587.2
ENST00000420859.1 ENST00000452294.1 ENST00000456894.1 |
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr6_-_119031228 | 0.03 |
ENST00000392500.3
ENST00000368488.5 ENST00000434604.1 |
CEP85L
|
centrosomal protein 85kDa-like |
| chr14_-_21493123 | 0.03 |
ENST00000556147.1
ENST00000554489.1 ENST00000555657.1 ENST00000557274.1 ENST00000555158.1 ENST00000554833.1 ENST00000555384.1 ENST00000556420.1 ENST00000554893.1 ENST00000553503.1 ENST00000555733.1 ENST00000553867.1 ENST00000397856.3 ENST00000397855.3 ENST00000556008.1 ENST00000557182.1 ENST00000554483.1 ENST00000556688.1 ENST00000397853.3 ENST00000556329.2 ENST00000554143.1 ENST00000397851.2 ENST00000555142.1 ENST00000557676.1 ENST00000556924.1 |
NDRG2
|
NDRG family member 2 |
| chr16_+_57220193 | 0.03 |
ENST00000564435.1
ENST00000562959.1 ENST00000394420.4 ENST00000568505.2 ENST00000537866.1 |
RSPRY1
|
ring finger and SPRY domain containing 1 |
| chr5_+_149887672 | 0.03 |
ENST00000261797.6
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr17_-_34122596 | 0.03 |
ENST00000250144.8
|
MMP28
|
matrix metallopeptidase 28 |
| chr20_-_50159198 | 0.03 |
ENST00000371564.3
ENST00000396009.3 ENST00000610033.1 |
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 |
| chr15_+_42841008 | 0.03 |
ENST00000260372.3
ENST00000568876.1 ENST00000568846.2 ENST00000562398.1 |
HAUS2
|
HAUS augmin-like complex, subunit 2 |
| chr2_-_113594279 | 0.03 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr7_+_23637118 | 0.03 |
ENST00000448353.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chr20_-_62601218 | 0.03 |
ENST00000369888.1
|
ZNF512B
|
zinc finger protein 512B |
| chr2_+_39005336 | 0.03 |
ENST00000409566.1
|
GEMIN6
|
gem (nuclear organelle) associated protein 6 |
| chr3_-_128880125 | 0.03 |
ENST00000393295.3
|
ISY1
|
ISY1 splicing factor homolog (S. cerevisiae) |
| chr9_+_125486269 | 0.03 |
ENST00000259466.1
|
OR1L4
|
olfactory receptor, family 1, subfamily L, member 4 |
| chr11_+_64052294 | 0.03 |
ENST00000536667.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr4_+_108911036 | 0.03 |
ENST00000505878.1
|
HADH
|
hydroxyacyl-CoA dehydrogenase |
| chr12_+_106751436 | 0.03 |
ENST00000228347.4
|
POLR3B
|
polymerase (RNA) III (DNA directed) polypeptide B |
| chr4_+_95972822 | 0.03 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr7_-_97501469 | 0.03 |
ENST00000414884.1
ENST00000442734.1 |
ASNS
|
asparagine synthetase (glutamine-hydrolyzing) |
| chr3_+_50388126 | 0.03 |
ENST00000425346.1
ENST00000424512.1 ENST00000232508.5 ENST00000418577.1 ENST00000606589.1 |
CYB561D2
XXcos-LUCA11.5
|
cytochrome b561 family, member D2 Uncharacterized protein |
| chr13_+_78109804 | 0.03 |
ENST00000535157.1
|
SCEL
|
sciellin |
| chr3_-_128879875 | 0.03 |
ENST00000418265.1
ENST00000393292.3 ENST00000273541.8 |
ISY1-RAB43
ISY1
|
ISY1-RAB43 readthrough ISY1 splicing factor homolog (S. cerevisiae) |
| chr20_-_50418972 | 0.03 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr6_+_33378517 | 0.03 |
ENST00000428274.1
|
PHF1
|
PHD finger protein 1 |
| chr1_-_51425772 | 0.03 |
ENST00000371778.4
|
FAF1
|
Fas (TNFRSF6) associated factor 1 |
| chr1_+_154966058 | 0.03 |
ENST00000392487.1
|
LENEP
|
lens epithelial protein |
| chr5_+_43192311 | 0.03 |
ENST00000326035.2
|
NIM1
|
NIM1 serine/threonine protein kinase |
| chr3_-_56809534 | 0.03 |
ENST00000497267.1
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TGIF1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.0 | 0.3 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.1 | GO:0008052 | sensory organ boundary specification(GO:0008052) formation of organ boundary(GO:0010160) taste bud development(GO:0061193) |
| 0.0 | 0.1 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.0 | 0.1 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) isoquinoline alkaloid metabolic process(GO:0033076) |
| 0.0 | 0.1 | GO:0070541 | response to platinum ion(GO:0070541) |
| 0.0 | 0.1 | GO:0050760 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.1 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.1 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.1 | GO:1901073 | N-acetylglucosamine biosynthetic process(GO:0006045) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.0 | 0.0 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.1 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.0 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.0 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004325 | ferrochelatase activity(GO:0004325) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.1 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.0 | 0.1 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0009384 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |