Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
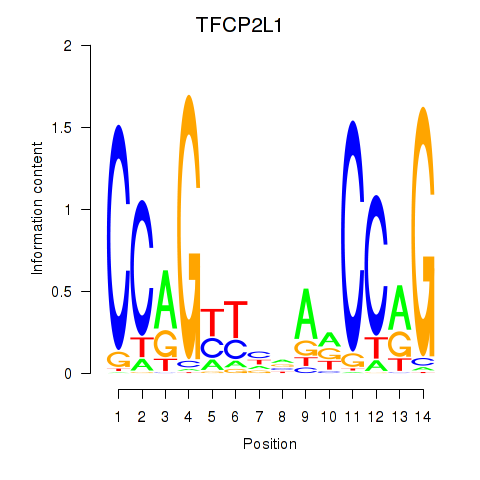
Results for TFCP2L1
Z-value: 0.66
Transcription factors associated with TFCP2L1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFCP2L1
|
ENSG00000115112.7 | transcription factor CP2 like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TFCP2L1 | hg19_v2_chr2_-_122042770_122042785 | 0.61 | 3.9e-01 | Click! |
Activity profile of TFCP2L1 motif
Sorted Z-values of TFCP2L1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_155959280 | 0.70 |
ENST00000495070.2
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr2_-_20212422 | 0.64 |
ENST00000421259.2
ENST00000407540.3 |
MATN3
|
matrilin 3 |
| chr19_-_36297632 | 0.41 |
ENST00000588266.2
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr1_+_27668505 | 0.39 |
ENST00000318074.5
|
SYTL1
|
synaptotagmin-like 1 |
| chr5_+_176513868 | 0.38 |
ENST00000292408.4
|
FGFR4
|
fibroblast growth factor receptor 4 |
| chr5_+_71475449 | 0.38 |
ENST00000504492.1
|
MAP1B
|
microtubule-associated protein 1B |
| chr6_-_31697977 | 0.37 |
ENST00000375787.2
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr22_-_20138302 | 0.36 |
ENST00000540078.1
ENST00000439765.2 |
AC006547.14
|
uncharacterized protein LOC388849 |
| chr3_+_52321827 | 0.34 |
ENST00000473032.1
ENST00000305690.8 ENST00000354773.4 ENST00000471180.1 ENST00000436784.2 |
GLYCTK
|
glycerate kinase |
| chr17_-_27038765 | 0.32 |
ENST00000581289.1
ENST00000301039.2 |
PROCA1
|
protein interacting with cyclin A1 |
| chrX_-_51239425 | 0.30 |
ENST00000375992.3
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr1_-_204329013 | 0.29 |
ENST00000272203.3
ENST00000414478.1 |
PLEKHA6
|
pleckstrin homology domain containing, family A member 6 |
| chr1_+_2477831 | 0.29 |
ENST00000606645.1
|
RP3-395M20.12
|
RP3-395M20.12 |
| chr1_-_207206092 | 0.29 |
ENST00000359470.5
ENST00000461135.2 |
C1orf116
|
chromosome 1 open reading frame 116 |
| chr4_-_122744998 | 0.28 |
ENST00000274026.5
|
CCNA2
|
cyclin A2 |
| chr5_+_176513895 | 0.28 |
ENST00000503708.1
ENST00000393648.2 ENST00000514472.1 ENST00000502906.1 ENST00000292410.3 ENST00000510911.1 |
FGFR4
|
fibroblast growth factor receptor 4 |
| chr11_-_17555421 | 0.28 |
ENST00000526181.1
|
USH1C
|
Usher syndrome 1C (autosomal recessive, severe) |
| chr3_-_52488048 | 0.28 |
ENST00000232975.3
|
TNNC1
|
troponin C type 1 (slow) |
| chr5_+_162864575 | 0.28 |
ENST00000512163.1
ENST00000393929.1 ENST00000340828.2 ENST00000511683.2 ENST00000510097.1 ENST00000511490.2 ENST00000510664.1 |
CCNG1
|
cyclin G1 |
| chr6_-_31550192 | 0.27 |
ENST00000429299.2
ENST00000446745.2 |
LTB
|
lymphotoxin beta (TNF superfamily, member 3) |
| chr17_-_27038063 | 0.26 |
ENST00000439862.3
|
PROCA1
|
protein interacting with cyclin A1 |
| chr4_-_164253738 | 0.26 |
ENST00000509586.1
ENST00000504391.1 ENST00000512819.1 |
NPY1R
|
neuropeptide Y receptor Y1 |
| chr2_-_61389240 | 0.25 |
ENST00000606876.1
|
RP11-493E12.1
|
RP11-493E12.1 |
| chr17_-_27467418 | 0.24 |
ENST00000528564.1
|
MYO18A
|
myosin XVIIIA |
| chr10_+_99344071 | 0.23 |
ENST00000370647.4
ENST00000370646.4 |
HOGA1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr12_-_124456598 | 0.22 |
ENST00000539761.1
ENST00000539551.1 |
CCDC92
|
coiled-coil domain containing 92 |
| chr2_+_103236004 | 0.22 |
ENST00000233969.2
|
SLC9A2
|
solute carrier family 9, subfamily A (NHE2, cation proton antiporter 2), member 2 |
| chr20_+_34680595 | 0.21 |
ENST00000406771.2
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr19_+_13988061 | 0.21 |
ENST00000339133.5
ENST00000397555.2 |
NANOS3
|
nanos homolog 3 (Drosophila) |
| chr12_+_56473939 | 0.21 |
ENST00000450146.2
|
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr15_+_76016293 | 0.21 |
ENST00000332145.2
|
ODF3L1
|
outer dense fiber of sperm tails 3-like 1 |
| chr15_-_62352570 | 0.21 |
ENST00000261517.5
ENST00000395896.4 ENST00000395898.3 |
VPS13C
|
vacuolar protein sorting 13 homolog C (S. cerevisiae) |
| chr19_+_46531127 | 0.21 |
ENST00000601033.1
|
CTC-344H19.4
|
CTC-344H19.4 |
| chr12_+_56473628 | 0.20 |
ENST00000549282.1
ENST00000549061.1 ENST00000267101.3 |
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr12_+_56473910 | 0.20 |
ENST00000411731.2
|
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr19_-_6481776 | 0.20 |
ENST00000543576.1
ENST00000590173.1 ENST00000381480.2 |
DENND1C
|
DENN/MADD domain containing 1C |
| chr2_+_219125714 | 0.20 |
ENST00000522678.1
ENST00000519574.1 ENST00000521462.1 |
GPBAR1
|
G protein-coupled bile acid receptor 1 |
| chr14_+_32546274 | 0.19 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr19_+_41860047 | 0.19 |
ENST00000604123.1
|
TMEM91
|
transmembrane protein 91 |
| chr19_+_41305612 | 0.19 |
ENST00000594380.1
ENST00000593397.1 ENST00000601733.1 |
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr12_+_14572070 | 0.19 |
ENST00000545769.1
ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr15_-_90234006 | 0.19 |
ENST00000300056.3
ENST00000559170.1 |
PEX11A
|
peroxisomal biogenesis factor 11 alpha |
| chr8_-_27850180 | 0.18 |
ENST00000380385.2
ENST00000301906.4 ENST00000354914.3 |
SCARA5
|
scavenger receptor class A, member 5 (putative) |
| chr22_+_40440804 | 0.18 |
ENST00000441751.1
ENST00000301923.9 |
TNRC6B
|
trinucleotide repeat containing 6B |
| chrX_-_106960285 | 0.18 |
ENST00000503515.1
ENST00000372397.2 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr19_+_41222998 | 0.18 |
ENST00000263370.2
|
ITPKC
|
inositol-trisphosphate 3-kinase C |
| chr19_+_19779589 | 0.17 |
ENST00000541458.1
|
ZNF101
|
zinc finger protein 101 |
| chr15_-_90233866 | 0.17 |
ENST00000561257.1
|
PEX11A
|
peroxisomal biogenesis factor 11 alpha |
| chr12_+_82752283 | 0.17 |
ENST00000548200.1
|
METTL25
|
methyltransferase like 25 |
| chr11_+_392587 | 0.17 |
ENST00000534401.1
|
PKP3
|
plakophilin 3 |
| chr6_-_30709980 | 0.16 |
ENST00000416018.1
ENST00000445853.1 ENST00000413165.1 ENST00000418160.1 |
FLOT1
|
flotillin 1 |
| chr20_-_62710832 | 0.16 |
ENST00000395042.1
|
RGS19
|
regulator of G-protein signaling 19 |
| chr17_+_75372165 | 0.15 |
ENST00000427674.2
|
SEPT9
|
septin 9 |
| chr5_+_60933634 | 0.15 |
ENST00000505642.1
|
C5orf64
|
chromosome 5 open reading frame 64 |
| chr5_+_95998714 | 0.15 |
ENST00000506811.1
ENST00000514055.1 |
CAST
|
calpastatin |
| chr11_+_65265141 | 0.15 |
ENST00000534336.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr12_+_57849048 | 0.15 |
ENST00000266646.2
|
INHBE
|
inhibin, beta E |
| chrY_+_2803322 | 0.15 |
ENST00000383052.1
ENST00000155093.3 ENST00000449237.1 ENST00000443793.1 |
ZFY
|
zinc finger protein, Y-linked |
| chr17_-_74023474 | 0.14 |
ENST00000301607.3
|
EVPL
|
envoplakin |
| chr20_+_34742650 | 0.14 |
ENST00000373945.1
ENST00000338074.2 |
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr14_-_61190754 | 0.14 |
ENST00000216513.4
|
SIX4
|
SIX homeobox 4 |
| chr4_-_76862117 | 0.14 |
ENST00000507956.1
ENST00000507187.2 ENST00000399497.3 ENST00000286733.4 |
NAAA
|
N-acylethanolamine acid amidase |
| chr19_+_24269981 | 0.14 |
ENST00000339642.6
ENST00000357002.4 |
ZNF254
|
zinc finger protein 254 |
| chr3_-_123411191 | 0.14 |
ENST00000354792.5
ENST00000508240.1 |
MYLK
|
myosin light chain kinase |
| chrX_-_63425561 | 0.14 |
ENST00000374869.3
ENST00000330258.3 |
AMER1
|
APC membrane recruitment protein 1 |
| chr12_-_124118151 | 0.14 |
ENST00000534960.1
|
EIF2B1
|
eukaryotic translation initiation factor 2B, subunit 1 alpha, 26kDa |
| chr10_-_75385711 | 0.14 |
ENST00000433394.1
|
USP54
|
ubiquitin specific peptidase 54 |
| chr19_-_12251130 | 0.14 |
ENST00000418866.1
ENST00000600335.1 |
ZNF20
|
zinc finger protein 20 |
| chr9_-_133769225 | 0.13 |
ENST00000343079.1
|
QRFP
|
pyroglutamylated RFamide peptide |
| chr17_+_55183261 | 0.13 |
ENST00000576295.1
|
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr19_+_11750566 | 0.13 |
ENST00000344893.3
|
ZNF833P
|
zinc finger protein 833, pseudogene |
| chr19_+_12075844 | 0.13 |
ENST00000592625.1
ENST00000586494.1 ENST00000343949.5 ENST00000545530.1 ENST00000358987.3 |
ZNF763
|
zinc finger protein 763 |
| chrX_+_24167746 | 0.13 |
ENST00000428571.1
ENST00000539115.1 |
ZFX
|
zinc finger protein, X-linked |
| chr11_+_18344106 | 0.13 |
ENST00000534641.1
ENST00000525831.1 ENST00000265963.4 |
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa |
| chr21_-_16437255 | 0.13 |
ENST00000400199.1
ENST00000400202.1 |
NRIP1
|
nuclear receptor interacting protein 1 |
| chr11_+_46383121 | 0.12 |
ENST00000454345.1
|
DGKZ
|
diacylglycerol kinase, zeta |
| chr6_-_30710265 | 0.12 |
ENST00000438162.1
ENST00000454845.1 |
FLOT1
|
flotillin 1 |
| chr19_-_16008880 | 0.12 |
ENST00000011989.7
ENST00000221700.6 |
CYP4F2
|
cytochrome P450, family 4, subfamily F, polypeptide 2 |
| chr17_+_1627834 | 0.12 |
ENST00000419248.1
ENST00000418841.1 |
WDR81
|
WD repeat domain 81 |
| chr4_-_174255536 | 0.12 |
ENST00000446922.2
|
HMGB2
|
high mobility group box 2 |
| chr1_-_45272951 | 0.12 |
ENST00000372200.1
|
TCTEX1D4
|
Tctex1 domain containing 4 |
| chr15_+_40226304 | 0.12 |
ENST00000559624.1
ENST00000382727.2 ENST00000263791.5 ENST00000560648.1 |
EIF2AK4
|
eukaryotic translation initiation factor 2 alpha kinase 4 |
| chr21_-_16437126 | 0.12 |
ENST00000318948.4
|
NRIP1
|
nuclear receptor interacting protein 1 |
| chr10_+_99344104 | 0.12 |
ENST00000555577.1
ENST00000370649.3 |
PI4K2A
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha Phosphatidylinositol 4-kinase type 2-alpha; Uncharacterized protein |
| chr9_+_101867387 | 0.11 |
ENST00000374990.2
ENST00000552516.1 |
TGFBR1
|
transforming growth factor, beta receptor 1 |
| chr4_-_987164 | 0.11 |
ENST00000398520.2
|
SLC26A1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr17_+_15848231 | 0.11 |
ENST00000304222.2
|
ADORA2B
|
adenosine A2b receptor |
| chr10_+_27793257 | 0.11 |
ENST00000375802.3
|
RAB18
|
RAB18, member RAS oncogene family |
| chr17_-_74023291 | 0.11 |
ENST00000586740.1
|
EVPL
|
envoplakin |
| chr14_+_24439148 | 0.11 |
ENST00000543805.1
ENST00000534993.1 |
DHRS4L2
|
dehydrogenase/reductase (SDR family) member 4 like 2 |
| chr19_-_6375860 | 0.11 |
ENST00000245810.1
|
PSPN
|
persephin |
| chr19_+_19779686 | 0.11 |
ENST00000415784.2
|
ZNF101
|
zinc finger protein 101 |
| chr11_+_46366918 | 0.11 |
ENST00000528615.1
ENST00000395574.3 |
DGKZ
|
diacylglycerol kinase, zeta |
| chr22_-_38484922 | 0.11 |
ENST00000428572.1
|
BAIAP2L2
|
BAI1-associated protein 2-like 2 |
| chr7_-_43965937 | 0.11 |
ENST00000455877.1
ENST00000223341.7 ENST00000447717.3 ENST00000426198.1 |
URGCP
|
upregulator of cell proliferation |
| chr17_+_40996590 | 0.10 |
ENST00000253799.3
ENST00000452774.2 |
AOC2
|
amine oxidase, copper containing 2 (retina-specific) |
| chr5_+_79703823 | 0.10 |
ENST00000338008.5
ENST00000510158.1 ENST00000505560.1 |
ZFYVE16
|
zinc finger, FYVE domain containing 16 |
| chr2_+_241544834 | 0.10 |
ENST00000319838.5
ENST00000403859.1 ENST00000438013.2 |
GPR35
|
G protein-coupled receptor 35 |
| chr11_+_73000449 | 0.10 |
ENST00000535931.1
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr11_-_77899634 | 0.10 |
ENST00000526208.1
ENST00000529350.1 ENST00000530018.1 ENST00000528776.1 |
KCTD21
|
potassium channel tetramerization domain containing 21 |
| chr10_+_27793197 | 0.10 |
ENST00000356940.6
ENST00000535776.1 |
RAB18
|
RAB18, member RAS oncogene family |
| chr11_-_31531121 | 0.10 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chrX_+_69674943 | 0.10 |
ENST00000542398.1
|
DLG3
|
discs, large homolog 3 (Drosophila) |
| chr1_+_110198689 | 0.10 |
ENST00000369836.4
|
GSTM4
|
glutathione S-transferase mu 4 |
| chr8_+_55370487 | 0.10 |
ENST00000297316.4
|
SOX17
|
SRY (sex determining region Y)-box 17 |
| chr1_-_1243252 | 0.10 |
ENST00000353662.3
|
ACAP3
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
| chr14_+_64971292 | 0.10 |
ENST00000358738.3
ENST00000394712.2 |
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr8_+_117778736 | 0.10 |
ENST00000309822.2
ENST00000357148.3 ENST00000517814.1 ENST00000517820.1 |
UTP23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chr17_+_75369167 | 0.10 |
ENST00000423034.2
|
SEPT9
|
septin 9 |
| chr10_+_35416223 | 0.10 |
ENST00000489321.1
ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM
|
cAMP responsive element modulator |
| chr19_+_41305330 | 0.09 |
ENST00000593972.1
|
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr9_+_91606355 | 0.09 |
ENST00000358157.2
|
S1PR3
|
sphingosine-1-phosphate receptor 3 |
| chr12_+_82752647 | 0.09 |
ENST00000550058.1
|
METTL25
|
methyltransferase like 25 |
| chr11_+_67776012 | 0.09 |
ENST00000539229.1
|
ALDH3B1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr1_+_174843548 | 0.09 |
ENST00000478442.1
ENST00000465412.1 |
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr7_+_73275483 | 0.09 |
ENST00000320531.2
|
WBSCR28
|
Williams-Beuren syndrome chromosome region 28 |
| chr8_-_103876965 | 0.09 |
ENST00000337198.5
|
AZIN1
|
antizyme inhibitor 1 |
| chrX_+_150565653 | 0.09 |
ENST00000330374.6
|
VMA21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr1_+_27189631 | 0.09 |
ENST00000339276.4
|
SFN
|
stratifin |
| chr19_+_41305627 | 0.09 |
ENST00000593525.1
|
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr11_+_62649158 | 0.09 |
ENST00000539891.1
ENST00000536981.1 |
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr11_-_119293903 | 0.09 |
ENST00000580275.1
|
THY1
|
Thy-1 cell surface antigen |
| chr19_+_41305406 | 0.09 |
ENST00000406058.2
ENST00000593726.1 |
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr16_+_333152 | 0.09 |
ENST00000219406.6
ENST00000404312.1 ENST00000456379.1 |
PDIA2
|
protein disulfide isomerase family A, member 2 |
| chr19_-_4338783 | 0.09 |
ENST00000601482.1
ENST00000600324.1 |
STAP2
|
signal transducing adaptor family member 2 |
| chr1_-_1356719 | 0.09 |
ENST00000520296.1
|
ANKRD65
|
ankyrin repeat domain 65 |
| chr5_+_95998746 | 0.09 |
ENST00000508608.1
|
CAST
|
calpastatin |
| chr12_-_110883346 | 0.09 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr12_+_124118366 | 0.09 |
ENST00000539994.1
ENST00000538845.1 ENST00000228955.7 ENST00000543341.2 ENST00000536375.1 |
GTF2H3
|
general transcription factor IIH, polypeptide 3, 34kDa |
| chr11_-_76155700 | 0.09 |
ENST00000572035.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr4_+_106629929 | 0.09 |
ENST00000512828.1
ENST00000394730.3 ENST00000507281.1 ENST00000515279.1 |
GSTCD
|
glutathione S-transferase, C-terminal domain containing |
| chr1_+_110198714 | 0.08 |
ENST00000336075.5
ENST00000326729.5 |
GSTM4
|
glutathione S-transferase mu 4 |
| chr2_-_233415220 | 0.08 |
ENST00000408957.3
|
TIGD1
|
tigger transposable element derived 1 |
| chrX_-_99891796 | 0.08 |
ENST00000373020.4
|
TSPAN6
|
tetraspanin 6 |
| chr11_-_119293872 | 0.08 |
ENST00000524970.1
|
THY1
|
Thy-1 cell surface antigen |
| chr1_-_11863571 | 0.08 |
ENST00000376583.3
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr3_+_184081137 | 0.08 |
ENST00000443489.1
|
POLR2H
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr10_+_123923205 | 0.08 |
ENST00000369004.3
ENST00000260733.3 |
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr1_-_1356628 | 0.08 |
ENST00000442470.1
ENST00000537107.1 |
ANKRD65
|
ankyrin repeat domain 65 |
| chr11_+_18343800 | 0.08 |
ENST00000453096.2
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa |
| chr12_+_82752275 | 0.08 |
ENST00000248306.3
|
METTL25
|
methyltransferase like 25 |
| chr8_-_54755459 | 0.08 |
ENST00000524234.1
ENST00000521275.1 ENST00000396774.2 |
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H |
| chr3_-_176915215 | 0.08 |
ENST00000457928.2
ENST00000422442.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr1_-_46089639 | 0.08 |
ENST00000445048.2
|
CCDC17
|
coiled-coil domain containing 17 |
| chr17_-_10633291 | 0.08 |
ENST00000578345.1
ENST00000455996.2 |
TMEM220
|
transmembrane protein 220 |
| chr2_-_31361543 | 0.08 |
ENST00000349752.5
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr10_+_123922941 | 0.08 |
ENST00000360561.3
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr5_-_151784838 | 0.08 |
ENST00000255262.3
|
NMUR2
|
neuromedin U receptor 2 |
| chr3_-_66551397 | 0.07 |
ENST00000383703.3
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr16_+_67562702 | 0.07 |
ENST00000379312.3
ENST00000042381.4 ENST00000540839.3 |
FAM65A
|
family with sequence similarity 65, member A |
| chr4_+_190992387 | 0.07 |
ENST00000554906.2
ENST00000553820.2 |
DUX4L7
|
double homeobox 4 like 7 |
| chr12_-_74686314 | 0.07 |
ENST00000551210.1
ENST00000515416.2 ENST00000549905.1 |
RP11-81H3.2
|
RP11-81H3.2 |
| chr19_-_4338838 | 0.07 |
ENST00000594605.1
|
STAP2
|
signal transducing adaptor family member 2 |
| chr16_+_46918235 | 0.07 |
ENST00000340124.4
|
GPT2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr6_+_84569359 | 0.07 |
ENST00000369681.5
ENST00000369679.4 |
CYB5R4
|
cytochrome b5 reductase 4 |
| chr9_+_71394945 | 0.07 |
ENST00000394264.3
|
FAM122A
|
family with sequence similarity 122A |
| chr3_-_122134882 | 0.07 |
ENST00000330689.4
|
WDR5B
|
WD repeat domain 5B |
| chr19_-_20844368 | 0.07 |
ENST00000595094.1
ENST00000601440.1 ENST00000291750.6 |
CTC-513N18.7
ZNF626
|
CTC-513N18.7 zinc finger protein 626 |
| chr1_-_89458604 | 0.07 |
ENST00000260508.4
|
CCBL2
|
cysteine conjugate-beta lyase 2 |
| chr3_+_121289551 | 0.07 |
ENST00000334384.3
|
ARGFX
|
arginine-fifty homeobox |
| chr9_+_108424738 | 0.07 |
ENST00000334077.3
|
TAL2
|
T-cell acute lymphocytic leukemia 2 |
| chr4_-_987217 | 0.07 |
ENST00000361661.2
ENST00000398516.2 |
SLC26A1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr5_+_95998673 | 0.07 |
ENST00000514845.1
|
CAST
|
calpastatin |
| chr7_-_140179276 | 0.07 |
ENST00000443720.2
ENST00000255977.2 |
MKRN1
|
makorin ring finger protein 1 |
| chr10_+_123923105 | 0.07 |
ENST00000368999.1
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr4_-_25865159 | 0.07 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr11_+_46366799 | 0.07 |
ENST00000532868.2
|
DGKZ
|
diacylglycerol kinase, zeta |
| chr9_+_35829208 | 0.07 |
ENST00000439587.2
ENST00000377991.4 |
TMEM8B
|
transmembrane protein 8B |
| chr11_+_96123158 | 0.07 |
ENST00000332349.4
ENST00000458427.1 |
JRKL
|
jerky homolog-like (mouse) |
| chr2_+_95873257 | 0.07 |
ENST00000425953.1
|
AC092835.2
|
AC092835.2 |
| chr6_-_52860171 | 0.07 |
ENST00000370963.4
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr11_+_65190245 | 0.06 |
ENST00000499732.1
ENST00000501122.2 ENST00000601801.1 |
NEAT1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
| chr7_-_64023410 | 0.06 |
ENST00000447137.2
|
ZNF680
|
zinc finger protein 680 |
| chr3_-_66551351 | 0.06 |
ENST00000273261.3
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr4_+_128982490 | 0.06 |
ENST00000394288.3
ENST00000432347.2 ENST00000264584.5 ENST00000441387.1 ENST00000427266.1 ENST00000354456.3 |
LARP1B
|
La ribonucleoprotein domain family, member 1B |
| chr19_-_9929484 | 0.06 |
ENST00000586651.1
ENST00000586073.1 |
FBXL12
|
F-box and leucine-rich repeat protein 12 |
| chr20_+_44509857 | 0.06 |
ENST00000372523.1
ENST00000372520.1 |
ZSWIM1
|
zinc finger, SWIM-type containing 1 |
| chr6_-_38607628 | 0.06 |
ENST00000498633.1
|
BTBD9
|
BTB (POZ) domain containing 9 |
| chr9_+_131451480 | 0.06 |
ENST00000322030.8
|
SET
|
SET nuclear oncogene |
| chr20_-_30458354 | 0.06 |
ENST00000428829.1
|
DUSP15
|
dual specificity phosphatase 15 |
| chr17_+_16318909 | 0.06 |
ENST00000577397.1
|
TRPV2
|
transient receptor potential cation channel, subfamily V, member 2 |
| chr2_-_27603582 | 0.06 |
ENST00000323703.6
ENST00000436006.1 |
ZNF513
|
zinc finger protein 513 |
| chr4_-_174255400 | 0.06 |
ENST00000506267.1
|
HMGB2
|
high mobility group box 2 |
| chr4_+_83351715 | 0.06 |
ENST00000273920.3
|
ENOPH1
|
enolase-phosphatase 1 |
| chr7_+_87563557 | 0.06 |
ENST00000439864.1
ENST00000412441.1 ENST00000398201.4 ENST00000265727.7 ENST00000315984.7 ENST00000398209.3 |
ADAM22
|
ADAM metallopeptidase domain 22 |
| chr7_-_64023441 | 0.06 |
ENST00000309683.6
|
ZNF680
|
zinc finger protein 680 |
| chr6_-_52859968 | 0.06 |
ENST00000370959.1
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr19_+_41305085 | 0.06 |
ENST00000303961.4
|
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr19_-_6591113 | 0.06 |
ENST00000423145.3
ENST00000245903.3 |
CD70
|
CD70 molecule |
| chr1_-_89458287 | 0.06 |
ENST00000370485.2
|
CCBL2
|
cysteine conjugate-beta lyase 2 |
| chr5_+_145583156 | 0.06 |
ENST00000265271.5
|
RBM27
|
RNA binding motif protein 27 |
| chr5_+_139739772 | 0.06 |
ENST00000506757.2
ENST00000230993.6 ENST00000506545.1 ENST00000432095.2 ENST00000507527.1 |
SLC4A9
|
solute carrier family 4, sodium bicarbonate cotransporter, member 9 |
| chr12_-_48499826 | 0.06 |
ENST00000551798.1
|
SENP1
|
SUMO1/sentrin specific peptidase 1 |
| chr5_-_36241900 | 0.06 |
ENST00000381937.4
ENST00000514504.1 |
NADK2
|
NAD kinase 2, mitochondrial |
| chr16_+_680932 | 0.05 |
ENST00000319070.2
|
WFIKKN1
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 1 |
| chr5_+_145583107 | 0.05 |
ENST00000506502.1
|
RBM27
|
RNA binding motif protein 27 |
| chr22_+_39898391 | 0.05 |
ENST00000434364.1
|
MIEF1
|
mitochondrial elongation factor 1 |
| chr5_-_169626104 | 0.05 |
ENST00000520275.1
ENST00000506431.2 |
CTB-27N1.1
|
CTB-27N1.1 |
| chr2_-_20189819 | 0.05 |
ENST00000281405.4
ENST00000345530.3 |
WDR35
|
WD repeat domain 35 |
| chr6_-_33679452 | 0.05 |
ENST00000374231.4
ENST00000607484.1 ENST00000374214.3 |
UQCC2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TFCP2L1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.1 | 0.7 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.1 | 0.3 | GO:0071373 | cellular response to cocaine(GO:0071314) cellular response to luteinizing hormone stimulus(GO:0071373) |
| 0.1 | 0.7 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 0.2 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.1 | 0.3 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.1 | 0.3 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.1 | 0.2 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.1 | 0.2 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.1 | 0.4 | GO:0010133 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.3 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.3 | GO:1901907 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.2 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.1 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.0 | 0.5 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.1 | GO:0071264 | regulation of eIF2 alpha phosphorylation by amino acid starvation(GO:0060733) regulation of translational initiation in response to starvation(GO:0071262) positive regulation of translational initiation in response to starvation(GO:0071264) |
| 0.0 | 0.1 | GO:0002276 | basophil activation involved in immune response(GO:0002276) basophil activation(GO:0045575) |
| 0.0 | 0.1 | GO:1905071 | occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.0 | 0.3 | GO:0044375 | peroxisome membrane biogenesis(GO:0016557) regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.1 | GO:0042662 | endodermal cell fate determination(GO:0007493) negative regulation of mesodermal cell fate specification(GO:0042662) regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) gall bladder development(GO:0061010) |
| 0.0 | 0.3 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.2 | GO:0038183 | bile acid signaling pathway(GO:0038183) |
| 0.0 | 0.1 | GO:0002880 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.1 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.1 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.6 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.0 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.2 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.2 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.0 | 0.2 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.2 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.4 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.4 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 | 0.1 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.3 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.0 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.5 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.2 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.3 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.1 | GO:0072244 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.0 | 0.1 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.2 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.4 | GO:1900153 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.3 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.0 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.0 | 0.3 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.1 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.1 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.3 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.3 | GO:0001602 | peptide YY receptor activity(GO:0001601) pancreatic polypeptide receptor activity(GO:0001602) |
| 0.1 | 0.7 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.4 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.5 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.3 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.1 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.0 | 0.6 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.1 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.3 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.0 | 0.2 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.2 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.0 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.2 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.0 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.0 | GO:0001002 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.6 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0045029 | uridine nucleotide receptor activity(GO:0015065) UDP-activated nucleotide receptor activity(GO:0045029) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |