Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
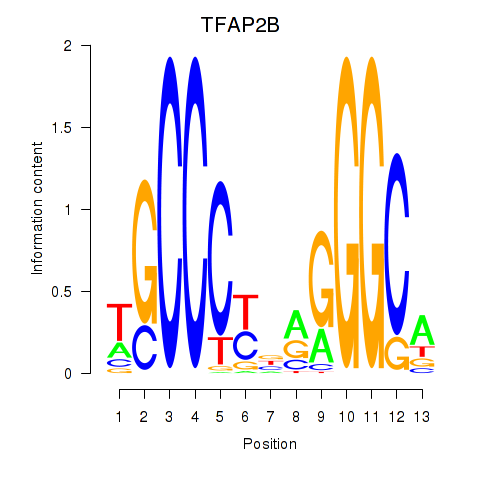
Results for TFAP2B
Z-value: 1.16
Transcription factors associated with TFAP2B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFAP2B
|
ENSG00000008196.8 | transcription factor AP-2 beta |
Activity profile of TFAP2B motif
Sorted Z-values of TFAP2B motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_19739321 | 0.78 |
ENST00000588461.1
|
LPAR2
|
lysophosphatidic acid receptor 2 |
| chr19_-_39832563 | 0.64 |
ENST00000599274.1
|
CTC-246B18.10
|
CTC-246B18.10 |
| chr17_+_36508826 | 0.42 |
ENST00000580660.1
|
SOCS7
|
suppressor of cytokine signaling 7 |
| chr17_+_7758374 | 0.41 |
ENST00000301599.6
ENST00000574668.1 |
TMEM88
|
transmembrane protein 88 |
| chr9_-_34523027 | 0.40 |
ENST00000399775.2
|
ENHO
|
energy homeostasis associated |
| chr2_-_55459485 | 0.40 |
ENST00000451916.1
|
CLHC1
|
clathrin heavy chain linker domain containing 1 |
| chr6_-_35888858 | 0.34 |
ENST00000507909.1
|
SRPK1
|
SRSF protein kinase 1 |
| chr1_+_221054411 | 0.34 |
ENST00000427693.1
|
HLX
|
H2.0-like homeobox |
| chr1_-_42921915 | 0.32 |
ENST00000372565.3
ENST00000433602.2 |
ZMYND12
|
zinc finger, MYND-type containing 12 |
| chr17_-_42767092 | 0.32 |
ENST00000588687.1
|
CCDC43
|
coiled-coil domain containing 43 |
| chr11_-_63684316 | 0.31 |
ENST00000301459.4
|
RCOR2
|
REST corepressor 2 |
| chr16_-_75498450 | 0.31 |
ENST00000566594.1
|
RP11-77K12.1
|
Uncharacterized protein |
| chr11_+_810221 | 0.29 |
ENST00000530398.1
|
RPLP2
|
ribosomal protein, large, P2 |
| chr6_+_33378517 | 0.27 |
ENST00000428274.1
|
PHF1
|
PHD finger protein 1 |
| chr4_+_1723197 | 0.26 |
ENST00000485989.2
ENST00000313288.4 |
TACC3
|
transforming, acidic coiled-coil containing protein 3 |
| chr22_-_31742218 | 0.26 |
ENST00000266269.5
ENST00000405309.3 ENST00000351933.4 |
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr11_-_64013663 | 0.26 |
ENST00000392210.2
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr3_+_158450143 | 0.25 |
ENST00000491804.1
|
MFSD1
|
major facilitator superfamily domain containing 1 |
| chr9_+_136325149 | 0.25 |
ENST00000542192.1
|
CACFD1
|
calcium channel flower domain containing 1 |
| chr19_-_55836697 | 0.25 |
ENST00000438693.1
ENST00000591570.1 |
TMEM150B
|
transmembrane protein 150B |
| chr8_-_103136481 | 0.24 |
ENST00000524209.1
ENST00000517822.1 ENST00000523923.1 ENST00000521599.1 ENST00000521964.1 ENST00000311028.3 ENST00000518166.1 |
NCALD
|
neurocalcin delta |
| chrX_+_131157322 | 0.23 |
ENST00000481105.1
ENST00000354719.6 ENST00000394335.2 |
MST4
|
Serine/threonine-protein kinase MST4 |
| chr15_+_75335604 | 0.23 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr19_+_19144384 | 0.23 |
ENST00000392335.2
ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6
|
armadillo repeat containing 6 |
| chr8_-_30670053 | 0.23 |
ENST00000518564.1
|
PPP2CB
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr4_-_1723040 | 0.22 |
ENST00000382936.3
ENST00000536901.1 ENST00000303277.2 |
TMEM129
|
transmembrane protein 129 |
| chr3_+_42544084 | 0.22 |
ENST00000543411.1
ENST00000438259.2 ENST00000439731.1 ENST00000325123.4 |
VIPR1
|
vasoactive intestinal peptide receptor 1 |
| chr5_+_60933634 | 0.22 |
ENST00000505642.1
|
C5orf64
|
chromosome 5 open reading frame 64 |
| chr16_-_66586365 | 0.22 |
ENST00000562484.2
|
TK2
|
thymidine kinase 2, mitochondrial |
| chr3_-_123168551 | 0.22 |
ENST00000462833.1
|
ADCY5
|
adenylate cyclase 5 |
| chr3_-_123710893 | 0.22 |
ENST00000467907.1
ENST00000459660.1 ENST00000495093.1 ENST00000460743.1 ENST00000405845.3 ENST00000484329.1 ENST00000479867.1 ENST00000496145.1 |
ROPN1
|
rhophilin associated tail protein 1 |
| chr14_+_65171315 | 0.22 |
ENST00000394691.1
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr13_-_107187462 | 0.21 |
ENST00000245323.4
|
EFNB2
|
ephrin-B2 |
| chr17_+_4613918 | 0.20 |
ENST00000574954.1
ENST00000346341.2 ENST00000572457.1 ENST00000381488.6 ENST00000412477.3 ENST00000571428.1 ENST00000575877.1 |
ARRB2
|
arrestin, beta 2 |
| chr19_-_55836669 | 0.20 |
ENST00000326652.4
|
TMEM150B
|
transmembrane protein 150B |
| chr2_-_85828867 | 0.20 |
ENST00000425160.1
|
TMEM150A
|
transmembrane protein 150A |
| chr3_-_49459878 | 0.20 |
ENST00000546031.1
ENST00000458307.2 ENST00000430521.1 |
AMT
|
aminomethyltransferase |
| chr3_-_138048631 | 0.20 |
ENST00000484930.1
ENST00000475751.1 |
NME9
|
NME/NM23 family member 9 |
| chr3_+_39424828 | 0.20 |
ENST00000273158.4
ENST00000431510.1 |
SLC25A38
|
solute carrier family 25, member 38 |
| chr11_-_535515 | 0.20 |
ENST00000311189.7
ENST00000451590.1 ENST00000417302.1 |
HRAS
|
Harvey rat sarcoma viral oncogene homolog |
| chr2_-_191399073 | 0.19 |
ENST00000421038.1
|
TMEM194B
|
transmembrane protein 194B |
| chr17_-_79869340 | 0.19 |
ENST00000538936.2
|
PCYT2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr14_+_65171099 | 0.19 |
ENST00000247226.7
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr3_-_49459865 | 0.19 |
ENST00000427987.1
|
AMT
|
aminomethyltransferase |
| chr17_-_61777459 | 0.19 |
ENST00000578993.1
ENST00000583211.1 ENST00000259006.3 |
LIMD2
|
LIM domain containing 2 |
| chr17_-_79869077 | 0.19 |
ENST00000570391.1
|
PCYT2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr19_-_17010360 | 0.19 |
ENST00000599287.2
|
CPAMD8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8 |
| chr17_-_72869086 | 0.19 |
ENST00000581530.1
ENST00000420580.2 ENST00000455107.2 ENST00000413947.2 ENST00000581219.1 ENST00000582944.1 |
FDXR
|
ferredoxin reductase |
| chr11_-_64013288 | 0.19 |
ENST00000542235.1
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr20_+_60174827 | 0.19 |
ENST00000543233.1
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal) |
| chr12_+_110011571 | 0.19 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr3_-_10052849 | 0.19 |
ENST00000437616.1
ENST00000429065.2 |
AC022007.5
|
AC022007.5 |
| chr16_+_67465016 | 0.18 |
ENST00000326152.5
|
HSD11B2
|
hydroxysteroid (11-beta) dehydrogenase 2 |
| chr11_+_64002292 | 0.18 |
ENST00000426086.2
|
VEGFB
|
vascular endothelial growth factor B |
| chr12_-_89919965 | 0.18 |
ENST00000548729.1
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough |
| chr17_-_79869004 | 0.18 |
ENST00000573927.1
ENST00000331285.3 ENST00000572157.1 |
PCYT2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr1_+_155179012 | 0.18 |
ENST00000609421.1
|
MTX1
|
metaxin 1 |
| chrX_-_108868390 | 0.18 |
ENST00000372101.2
|
KCNE1L
|
KCNE1-like |
| chrX_+_131157290 | 0.18 |
ENST00000394334.2
|
MST4
|
Serine/threonine-protein kinase MST4 |
| chr11_+_63974135 | 0.18 |
ENST00000544997.1
ENST00000345728.5 ENST00000279227.5 |
FERMT3
|
fermitin family member 3 |
| chr17_-_5138099 | 0.17 |
ENST00000571800.1
ENST00000574081.1 ENST00000399600.4 ENST00000574297.1 |
SCIMP
|
SLP adaptor and CSK interacting membrane protein |
| chr20_-_31124186 | 0.17 |
ENST00000375678.3
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr11_+_809961 | 0.17 |
ENST00000530797.1
|
RPLP2
|
ribosomal protein, large, P2 |
| chr19_-_39402798 | 0.17 |
ENST00000571838.1
|
CTC-360G5.1
|
coiled-coil glutamate-rich protein 2 |
| chr12_+_56414851 | 0.17 |
ENST00000547167.1
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr17_-_27230035 | 0.17 |
ENST00000378895.4
ENST00000394901.3 |
DHRS13
|
dehydrogenase/reductase (SDR family) member 13 |
| chr8_+_22462532 | 0.17 |
ENST00000389279.3
|
CCAR2
|
cell cycle and apoptosis regulator 2 |
| chr6_-_34664612 | 0.17 |
ENST00000374023.3
ENST00000374026.3 |
C6orf106
|
chromosome 6 open reading frame 106 |
| chr1_-_15911510 | 0.17 |
ENST00000375826.3
|
AGMAT
|
agmatine ureohydrolase (agmatinase) |
| chr22_+_31742875 | 0.17 |
ENST00000504184.2
|
AC005003.1
|
CDNA FLJ20464 fis, clone KAT06158; HCG1777549; Uncharacterized protein |
| chr10_+_104178946 | 0.17 |
ENST00000432590.1
|
FBXL15
|
F-box and leucine-rich repeat protein 15 |
| chr10_+_48355024 | 0.17 |
ENST00000395702.2
ENST00000442001.1 ENST00000433077.1 ENST00000436850.1 ENST00000494156.1 ENST00000586537.1 |
ZNF488
|
zinc finger protein 488 |
| chr19_-_14586125 | 0.17 |
ENST00000292513.3
|
PTGER1
|
prostaglandin E receptor 1 (subtype EP1), 42kDa |
| chr11_-_61348292 | 0.16 |
ENST00000539008.1
ENST00000540677.1 ENST00000542836.1 ENST00000542670.1 ENST00000535826.1 ENST00000545053.1 |
SYT7
|
synaptotagmin VII |
| chr8_+_22462145 | 0.16 |
ENST00000308511.4
ENST00000523801.1 ENST00000521301.1 |
CCAR2
|
cell cycle and apoptosis regulator 2 |
| chr22_-_30661807 | 0.16 |
ENST00000403389.1
|
OSM
|
oncostatin M |
| chr17_+_1627834 | 0.16 |
ENST00000419248.1
ENST00000418841.1 |
WDR81
|
WD repeat domain 81 |
| chr10_+_103825080 | 0.16 |
ENST00000299238.5
|
HPS6
|
Hermansky-Pudlak syndrome 6 |
| chr1_+_46269248 | 0.16 |
ENST00000361297.2
ENST00000372009.2 |
MAST2
|
microtubule associated serine/threonine kinase 2 |
| chr13_+_21714913 | 0.16 |
ENST00000450573.1
ENST00000467636.1 |
SAP18
|
Sin3A-associated protein, 18kDa |
| chr17_-_7760779 | 0.16 |
ENST00000335155.5
ENST00000575071.1 |
LSMD1
|
LSM domain containing 1 |
| chr3_+_107244229 | 0.16 |
ENST00000456419.1
ENST00000402163.2 |
BBX
|
bobby sox homolog (Drosophila) |
| chr12_+_113495492 | 0.16 |
ENST00000257600.3
|
DTX1
|
deltex homolog 1 (Drosophila) |
| chr5_+_141016969 | 0.16 |
ENST00000518856.1
|
RELL2
|
RELT-like 2 |
| chr12_-_57443886 | 0.16 |
ENST00000300119.3
|
MYO1A
|
myosin IA |
| chr17_-_72869140 | 0.16 |
ENST00000583917.1
ENST00000293195.5 ENST00000442102.2 |
FDXR
|
ferredoxin reductase |
| chr17_+_36861735 | 0.15 |
ENST00000378137.5
ENST00000325718.7 |
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chr20_+_44746885 | 0.15 |
ENST00000372285.3
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5 |
| chrX_-_150067272 | 0.15 |
ENST00000355149.3
ENST00000437787.2 |
CD99L2
|
CD99 molecule-like 2 |
| chr11_+_3877412 | 0.15 |
ENST00000527651.1
|
STIM1
|
stromal interaction molecule 1 |
| chrX_-_150067173 | 0.15 |
ENST00000370377.3
ENST00000320893.6 |
CD99L2
|
CD99 molecule-like 2 |
| chr1_-_44497118 | 0.15 |
ENST00000537678.1
ENST00000466926.1 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr7_+_16793160 | 0.15 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr3_+_10857885 | 0.15 |
ENST00000254488.2
ENST00000454147.1 |
SLC6A11
|
solute carrier family 6 (neurotransmitter transporter), member 11 |
| chr17_-_79869243 | 0.15 |
ENST00000538721.2
ENST00000573636.2 ENST00000571105.1 ENST00000576343.1 ENST00000572473.1 |
PCYT2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr5_-_141338377 | 0.15 |
ENST00000510041.1
|
PCDH12
|
protocadherin 12 |
| chr10_-_135238076 | 0.15 |
ENST00000414069.2
|
SPRN
|
shadow of prion protein homolog (zebrafish) |
| chr7_+_65939543 | 0.15 |
ENST00000600021.1
|
AC008267.1
|
HCG1983814; Uncharacterized protein |
| chr1_+_33722080 | 0.15 |
ENST00000483388.1
ENST00000539719.1 |
ZNF362
|
zinc finger protein 362 |
| chr16_-_67970990 | 0.15 |
ENST00000358514.4
|
PSMB10
|
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr8_-_145559943 | 0.14 |
ENST00000332135.4
|
SCRT1
|
scratch family zinc finger 1 |
| chr17_+_7748233 | 0.14 |
ENST00000570632.1
|
KDM6B
|
lysine (K)-specific demethylase 6B |
| chr19_+_14544099 | 0.14 |
ENST00000242783.6
ENST00000586557.1 ENST00000590097.1 |
PKN1
|
protein kinase N1 |
| chr11_-_65655906 | 0.14 |
ENST00000533045.1
ENST00000338369.2 ENST00000357519.4 |
FIBP
|
fibroblast growth factor (acidic) intracellular binding protein |
| chr6_-_41040049 | 0.14 |
ENST00000471367.1
|
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr11_+_64001962 | 0.14 |
ENST00000309422.2
|
VEGFB
|
vascular endothelial growth factor B |
| chr1_-_40782938 | 0.14 |
ENST00000372736.3
ENST00000372748.3 |
COL9A2
|
collagen, type IX, alpha 2 |
| chrX_-_154033686 | 0.14 |
ENST00000453245.1
ENST00000428488.1 ENST00000369531.1 |
MPP1
|
membrane protein, palmitoylated 1, 55kDa |
| chr19_+_7710774 | 0.14 |
ENST00000602355.1
|
STXBP2
|
syntaxin binding protein 2 |
| chr17_-_42992856 | 0.14 |
ENST00000588316.1
ENST00000435360.2 ENST00000586793.1 ENST00000588735.1 ENST00000588037.1 ENST00000592320.1 ENST00000253408.5 |
GFAP
|
glial fibrillary acidic protein |
| chr1_-_6321035 | 0.14 |
ENST00000377893.2
|
GPR153
|
G protein-coupled receptor 153 |
| chr1_-_49242553 | 0.14 |
ENST00000371833.3
|
BEND5
|
BEN domain containing 5 |
| chr9_+_136325089 | 0.14 |
ENST00000291722.7
ENST00000316948.4 ENST00000540581.1 |
CACFD1
|
calcium channel flower domain containing 1 |
| chr19_+_50706866 | 0.14 |
ENST00000440075.2
ENST00000376970.2 ENST00000425460.1 ENST00000599920.1 ENST00000601313.1 |
MYH14
|
myosin, heavy chain 14, non-muscle |
| chrX_-_19905577 | 0.14 |
ENST00000379697.3
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr10_-_8095412 | 0.14 |
ENST00000458727.1
ENST00000355358.1 |
RP11-379F12.3
GATA3-AS1
|
RP11-379F12.3 GATA3 antisense RNA 1 |
| chr12_+_7060414 | 0.13 |
ENST00000538715.1
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr16_+_88772866 | 0.13 |
ENST00000453996.2
ENST00000312060.5 ENST00000378384.3 ENST00000567949.1 ENST00000564921.1 |
CTU2
|
cytosolic thiouridylase subunit 2 homolog (S. pombe) |
| chr17_-_8093471 | 0.13 |
ENST00000389017.4
|
C17orf59
|
chromosome 17 open reading frame 59 |
| chr20_+_42965626 | 0.13 |
ENST00000217043.2
|
R3HDML
|
R3H domain containing-like |
| chr8_-_144654918 | 0.13 |
ENST00000529971.1
|
MROH6
|
maestro heat-like repeat family member 6 |
| chr9_+_108424738 | 0.13 |
ENST00000334077.3
|
TAL2
|
T-cell acute lymphocytic leukemia 2 |
| chr1_-_21948906 | 0.13 |
ENST00000374761.2
ENST00000599760.1 |
RAP1GAP
|
RAP1 GTPase activating protein |
| chr19_+_17858547 | 0.13 |
ENST00000600676.1
ENST00000600209.1 ENST00000596309.1 ENST00000598539.1 ENST00000597474.1 ENST00000593385.1 ENST00000598067.1 ENST00000593833.1 |
FCHO1
|
FCH domain only 1 |
| chr1_+_100818156 | 0.13 |
ENST00000336454.3
|
CDC14A
|
cell division cycle 14A |
| chr5_+_172068232 | 0.13 |
ENST00000520919.1
ENST00000522853.1 ENST00000369800.5 |
NEURL1B
|
neuralized E3 ubiquitin protein ligase 1B |
| chr17_-_79869228 | 0.13 |
ENST00000570388.1
|
PCYT2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr20_+_44746939 | 0.13 |
ENST00000372276.3
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5 |
| chr14_+_24540154 | 0.13 |
ENST00000559778.1
ENST00000560761.1 ENST00000557889.1 |
CPNE6
|
copine VI (neuronal) |
| chr12_+_56414795 | 0.13 |
ENST00000431367.2
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr13_+_99853124 | 0.13 |
ENST00000376440.2
ENST00000457666.1 |
UBAC2
|
UBA domain containing 2 |
| chr19_-_10450328 | 0.12 |
ENST00000160262.5
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr1_+_1447517 | 0.12 |
ENST00000378756.3
ENST00000378755.5 |
ATAD3A
|
ATPase family, AAA domain containing 3A |
| chr6_-_132272504 | 0.12 |
ENST00000367976.3
|
CTGF
|
connective tissue growth factor |
| chr3_-_185216766 | 0.12 |
ENST00000296254.3
|
TMEM41A
|
transmembrane protein 41A |
| chr19_-_49149553 | 0.12 |
ENST00000084798.4
|
CA11
|
carbonic anhydrase XI |
| chr5_-_176730676 | 0.12 |
ENST00000393611.2
ENST00000303251.6 ENST00000303270.6 |
RAB24
|
RAB24, member RAS oncogene family |
| chr16_-_11836595 | 0.12 |
ENST00000356957.3
ENST00000283033.5 |
TXNDC11
|
thioredoxin domain containing 11 |
| chr11_+_62379194 | 0.12 |
ENST00000525801.1
ENST00000534093.1 |
ROM1
|
retinal outer segment membrane protein 1 |
| chr17_-_39677971 | 0.12 |
ENST00000393976.2
|
KRT15
|
keratin 15 |
| chr1_+_155051379 | 0.12 |
ENST00000418360.2
|
EFNA3
|
ephrin-A3 |
| chr19_-_42724261 | 0.12 |
ENST00000595337.1
|
DEDD2
|
death effector domain containing 2 |
| chr3_-_183145765 | 0.12 |
ENST00000473233.1
|
MCF2L2
|
MCF.2 cell line derived transforming sequence-like 2 |
| chr17_-_4900873 | 0.12 |
ENST00000355025.3
ENST00000575780.1 ENST00000396829.2 |
INCA1
|
inhibitor of CDK, cyclin A1 interacting protein 1 |
| chr4_+_108911036 | 0.12 |
ENST00000505878.1
|
HADH
|
hydroxyacyl-CoA dehydrogenase |
| chr1_+_11994715 | 0.12 |
ENST00000449038.1
ENST00000376369.3 ENST00000429000.2 ENST00000196061.4 |
PLOD1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr17_-_36906058 | 0.12 |
ENST00000580830.1
|
PCGF2
|
polycomb group ring finger 2 |
| chr3_+_184097905 | 0.12 |
ENST00000450923.1
|
CHRD
|
chordin |
| chr17_-_41856305 | 0.11 |
ENST00000397937.2
ENST00000226004.3 |
DUSP3
|
dual specificity phosphatase 3 |
| chr19_-_10450287 | 0.11 |
ENST00000589261.1
ENST00000590569.1 ENST00000589580.1 ENST00000589249.1 |
ICAM3
|
intercellular adhesion molecule 3 |
| chr15_-_77363375 | 0.11 |
ENST00000559494.1
|
TSPAN3
|
tetraspanin 3 |
| chr19_+_38397839 | 0.11 |
ENST00000222345.6
|
SIPA1L3
|
signal-induced proliferation-associated 1 like 3 |
| chr1_+_153746683 | 0.11 |
ENST00000271857.2
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr6_+_43112037 | 0.11 |
ENST00000473339.1
|
PTK7
|
protein tyrosine kinase 7 |
| chr3_+_75713481 | 0.11 |
ENST00000308062.3
ENST00000464571.1 |
FRG2C
|
FSHD region gene 2 family, member C |
| chr16_-_28503080 | 0.11 |
ENST00000565316.1
ENST00000565778.1 ENST00000357857.9 ENST00000568558.1 ENST00000357806.7 |
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr1_+_155006300 | 0.11 |
ENST00000295542.1
ENST00000392480.1 ENST00000423025.2 ENST00000368419.2 |
DCST1
|
DC-STAMP domain containing 1 |
| chr1_+_100818009 | 0.11 |
ENST00000370125.2
ENST00000361544.6 ENST00000370124.3 |
CDC14A
|
cell division cycle 14A |
| chr4_+_108910870 | 0.11 |
ENST00000403312.1
ENST00000603302.1 ENST00000309522.3 |
HADH
|
hydroxyacyl-CoA dehydrogenase |
| chr7_+_128828713 | 0.11 |
ENST00000249373.3
|
SMO
|
smoothened, frizzled family receptor |
| chr17_+_59529743 | 0.11 |
ENST00000589003.1
ENST00000393853.4 |
TBX4
|
T-box 4 |
| chr6_-_31864977 | 0.11 |
ENST00000395728.3
ENST00000375528.4 |
EHMT2
|
euchromatic histone-lysine N-methyltransferase 2 |
| chr14_+_94640633 | 0.11 |
ENST00000304338.3
|
PPP4R4
|
protein phosphatase 4, regulatory subunit 4 |
| chr4_+_8271471 | 0.11 |
ENST00000307358.2
ENST00000382512.3 |
HTRA3
|
HtrA serine peptidase 3 |
| chr12_+_53693466 | 0.11 |
ENST00000267103.5
ENST00000548632.1 |
C12orf10
|
chromosome 12 open reading frame 10 |
| chr17_+_66511224 | 0.11 |
ENST00000588178.1
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr16_-_28223166 | 0.10 |
ENST00000304658.5
|
XPO6
|
exportin 6 |
| chr1_-_161039647 | 0.10 |
ENST00000368013.3
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr12_-_32908809 | 0.10 |
ENST00000324868.8
|
YARS2
|
tyrosyl-tRNA synthetase 2, mitochondrial |
| chr14_+_24458123 | 0.10 |
ENST00000545240.1
ENST00000382755.4 |
DHRS4L2
|
dehydrogenase/reductase (SDR family) member 4 like 2 |
| chr1_+_155178481 | 0.10 |
ENST00000368376.3
|
MTX1
|
metaxin 1 |
| chr19_-_55660561 | 0.10 |
ENST00000587758.1
ENST00000356783.5 ENST00000291901.8 ENST00000588426.1 ENST00000588147.1 ENST00000536926.1 ENST00000588981.1 |
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr11_-_64014379 | 0.10 |
ENST00000309318.3
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr7_+_127292234 | 0.10 |
ENST00000354725.3
|
SND1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr14_+_24458021 | 0.10 |
ENST00000397071.1
ENST00000559411.1 ENST00000335125.6 |
DHRS4L2
|
dehydrogenase/reductase (SDR family) member 4 like 2 |
| chr8_-_144655141 | 0.10 |
ENST00000398882.3
|
MROH6
|
maestro heat-like repeat family member 6 |
| chr1_-_6260896 | 0.10 |
ENST00000497965.1
|
RPL22
|
ribosomal protein L22 |
| chr1_-_161039753 | 0.10 |
ENST00000368015.1
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr12_+_6493199 | 0.10 |
ENST00000228918.4
|
LTBR
|
lymphotoxin beta receptor (TNFR superfamily, member 3) |
| chr20_+_44462749 | 0.10 |
ENST00000372541.1
|
SNX21
|
sorting nexin family member 21 |
| chr17_+_79953310 | 0.10 |
ENST00000582355.2
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr2_-_86564776 | 0.10 |
ENST00000165698.5
ENST00000541910.1 ENST00000535845.1 |
REEP1
|
receptor accessory protein 1 |
| chr5_-_176981417 | 0.10 |
ENST00000514747.1
ENST00000443375.2 ENST00000329540.5 |
FAM193B
|
family with sequence similarity 193, member B |
| chr5_-_93077293 | 0.10 |
ENST00000510627.4
|
POU5F2
|
POU domain class 5, transcription factor 2 |
| chr7_+_30811004 | 0.10 |
ENST00000265299.6
|
FAM188B
|
family with sequence similarity 188, member B |
| chr16_+_24857552 | 0.10 |
ENST00000568579.1
ENST00000567758.1 ENST00000569071.1 ENST00000539472.1 |
SLC5A11
|
solute carrier family 5 (sodium/inositol cotransporter), member 11 |
| chr22_+_46692638 | 0.10 |
ENST00000454366.1
|
GTSE1
|
G-2 and S-phase expressed 1 |
| chr7_+_76139741 | 0.10 |
ENST00000334348.3
ENST00000419923.2 ENST00000448265.3 ENST00000443097.2 |
UPK3B
|
uroplakin 3B |
| chr17_+_61678225 | 0.09 |
ENST00000258975.6
|
TACO1
|
translational activator of mitochondrially encoded cytochrome c oxidase I |
| chr12_+_49212514 | 0.09 |
ENST00000301050.2
ENST00000548279.1 ENST00000547230.1 |
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit |
| chr2_+_55459495 | 0.09 |
ENST00000272317.6
ENST00000449323.1 |
RPS27A
|
ribosomal protein S27a |
| chr16_-_66959429 | 0.09 |
ENST00000420652.1
ENST00000299759.6 |
RRAD
|
Ras-related associated with diabetes |
| chr7_+_150549565 | 0.09 |
ENST00000360937.4
ENST00000416793.2 ENST00000483043.1 |
AOC1
|
amine oxidase, copper containing 1 |
| chr9_-_131534160 | 0.09 |
ENST00000291900.2
|
ZER1
|
zyg-11 related, cell cycle regulator |
| chr19_+_42387228 | 0.09 |
ENST00000354532.3
ENST00000599846.1 ENST00000347545.4 |
ARHGEF1
|
Rho guanine nucleotide exchange factor (GEF) 1 |
| chr9_-_123639600 | 0.09 |
ENST00000373896.3
|
PHF19
|
PHD finger protein 19 |
| chr17_-_27405875 | 0.09 |
ENST00000359450.6
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr16_+_31085714 | 0.09 |
ENST00000300850.5
ENST00000564189.1 ENST00000428260.1 |
ZNF646
|
zinc finger protein 646 |
| chr17_-_4871085 | 0.09 |
ENST00000575142.1
ENST00000206020.3 |
SPAG7
|
sperm associated antigen 7 |
| chr1_+_172628154 | 0.09 |
ENST00000340030.3
ENST00000367721.2 |
FASLG
|
Fas ligand (TNF superfamily, member 6) |
Network of associatons between targets according to the STRING database.
First level regulatory network of TFAP2B
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 0.4 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.2 | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning(GO:0021919) |
| 0.0 | 0.3 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.2 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.0 | 0.1 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.0 | 0.1 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.0 | 0.2 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.1 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.3 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.3 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:1902910 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.0 | 0.3 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.2 | GO:0010629 | negative regulation of gene expression(GO:0010629) |
| 0.0 | 0.3 | GO:0033590 | response to cobalamin(GO:0033590) cellular response to erythropoietin(GO:0036018) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.2 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:0072313 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.0 | 0.4 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:0061145 | bronchus cartilage development(GO:0060532) lung smooth muscle development(GO:0061145) |
| 0.0 | 0.1 | GO:0035283 | rhombomere 3 development(GO:0021569) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.0 | 0.1 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.0 | 0.2 | GO:1901162 | primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.2 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.2 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.8 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.1 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.1 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.0 | 0.2 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.1 | GO:0035668 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.0 | 0.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.3 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.1 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.2 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.8 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.1 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.0 | 0.1 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:1904204 | skeletal muscle hypertrophy(GO:0014734) regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.1 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.1 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.0 | 0.1 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.0 | 0.1 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.0 | 0.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.2 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.1 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.1 | GO:0070309 | positive regulation by virus of viral protein levels in host cell(GO:0046726) lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.1 | GO:1901911 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.1 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.1 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.2 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.1 | GO:0048388 | cellular chloride ion homeostasis(GO:0030644) retinal cell programmed cell death(GO:0046666) endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.2 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.1 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.0 | 0.2 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.0 | 0.1 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.0 | 0.2 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.1 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 0.0 | 0.3 | GO:0051255 | spindle midzone assembly(GO:0051255) |
| 0.0 | 0.1 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.0 | GO:0002818 | intracellular defense response(GO:0002818) |
| 0.0 | 0.1 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.3 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.1 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.1 | GO:0003072 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.0 | 0.0 | GO:0070666 | mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.0 | GO:2001176 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.0 | 0.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.0 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.0 | 0.1 | GO:0044856 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.1 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.0 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.0 | 0.0 | GO:2000657 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.2 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.1 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.0 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.2 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.3 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.0 | GO:0097444 | spine apparatus(GO:0097444) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.1 | 0.3 | GO:0004324 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.1 | 0.4 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.1 | 0.8 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.2 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.1 | 0.2 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.2 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.2 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.1 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.0 | 0.1 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.2 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.1 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.0 | 0.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.1 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.1 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.1 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0010736 | serum response element binding(GO:0010736) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0052846 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.1 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.2 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.0 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.1 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |