Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
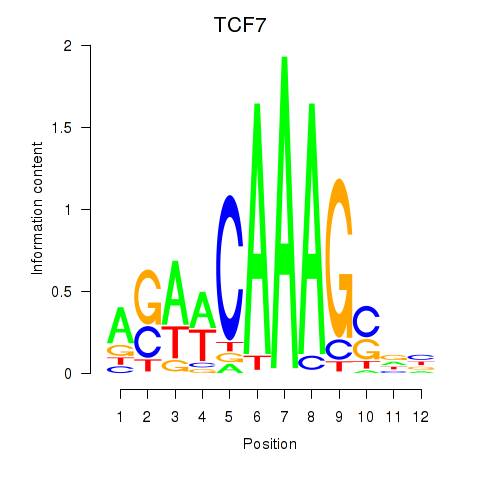
Results for TCF7
Z-value: 0.21
Transcription factors associated with TCF7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TCF7
|
ENSG00000081059.15 | transcription factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TCF7 | hg19_v2_chr5_+_133450365_133450444 | -0.57 | 4.3e-01 | Click! |
Activity profile of TCF7 motif
Sorted Z-values of TCF7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_32414059 | 0.16 |
ENST00000553330.1
|
RP11-187E13.1
|
Uncharacterized protein |
| chr20_+_57875457 | 0.12 |
ENST00000337938.2
ENST00000311585.7 ENST00000371028.2 |
EDN3
|
endothelin 3 |
| chr1_-_151345159 | 0.12 |
ENST00000458566.1
ENST00000447402.3 ENST00000426705.2 ENST00000435071.1 ENST00000368868.5 |
SELENBP1
|
selenium binding protein 1 |
| chr7_-_16840820 | 0.11 |
ENST00000450569.1
|
AGR2
|
anterior gradient 2 |
| chr17_-_20946710 | 0.11 |
ENST00000584538.1
|
USP22
|
ubiquitin specific peptidase 22 |
| chr3_-_167813672 | 0.11 |
ENST00000470487.1
|
GOLIM4
|
golgi integral membrane protein 4 |
| chr4_+_71859156 | 0.11 |
ENST00000286648.5
ENST00000504730.1 ENST00000504952.1 |
DCK
|
deoxycytidine kinase |
| chr17_+_68100989 | 0.11 |
ENST00000585558.1
ENST00000392670.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr17_-_57229155 | 0.10 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr4_+_95128748 | 0.10 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr10_+_54074033 | 0.10 |
ENST00000373970.3
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr12_+_21654714 | 0.10 |
ENST00000542038.1
ENST00000540141.1 ENST00000229314.5 |
GOLT1B
|
golgi transport 1B |
| chr12_+_100594557 | 0.10 |
ENST00000546902.1
ENST00000552376.1 ENST00000551617.1 |
ACTR6
|
ARP6 actin-related protein 6 homolog (yeast) |
| chr4_+_89300158 | 0.10 |
ENST00000502870.1
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr1_-_145470383 | 0.09 |
ENST00000369314.1
ENST00000369313.3 |
POLR3GL
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD)-like |
| chr22_-_37880543 | 0.09 |
ENST00000442496.1
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr4_-_139163491 | 0.09 |
ENST00000280612.5
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr10_-_25241499 | 0.09 |
ENST00000376378.1
ENST00000376376.3 ENST00000320152.6 |
PRTFDC1
|
phosphoribosyl transferase domain containing 1 |
| chr2_+_70056762 | 0.09 |
ENST00000282570.3
|
GMCL1
|
germ cell-less, spermatogenesis associated 1 |
| chr4_-_140477928 | 0.09 |
ENST00000274031.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr14_+_74486043 | 0.09 |
ENST00000464394.1
ENST00000394009.3 |
CCDC176
|
coiled-coil domain containing 176 |
| chr9_+_91003271 | 0.09 |
ENST00000375859.3
ENST00000541629.1 |
SPIN1
|
spindlin 1 |
| chr12_+_69080734 | 0.09 |
ENST00000378905.2
|
NUP107
|
nucleoporin 107kDa |
| chr17_+_34848049 | 0.09 |
ENST00000588902.1
ENST00000591067.1 |
ZNHIT3
|
zinc finger, HIT-type containing 3 |
| chr3_+_118892411 | 0.09 |
ENST00000479520.1
ENST00000494855.1 |
UPK1B
|
uroplakin 1B |
| chr7_+_99613195 | 0.08 |
ENST00000324306.6
|
ZKSCAN1
|
zinc finger with KRAB and SCAN domains 1 |
| chr6_+_43968306 | 0.08 |
ENST00000442114.2
ENST00000336600.5 ENST00000439969.2 |
C6orf223
|
chromosome 6 open reading frame 223 |
| chr10_+_99332198 | 0.08 |
ENST00000307518.5
ENST00000298808.5 ENST00000370655.1 |
ANKRD2
|
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr12_+_104359614 | 0.08 |
ENST00000266775.9
ENST00000544861.1 |
TDG
|
thymine-DNA glycosylase |
| chr3_+_118892362 | 0.08 |
ENST00000497685.1
ENST00000264234.3 |
UPK1B
|
uroplakin 1B |
| chr15_+_36871983 | 0.08 |
ENST00000437989.2
ENST00000569302.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr2_-_160761179 | 0.08 |
ENST00000554112.1
ENST00000553424.1 ENST00000263636.4 ENST00000504764.1 ENST00000505052.1 |
LY75
LY75-CD302
|
lymphocyte antigen 75 LY75-CD302 readthrough |
| chr6_+_64281906 | 0.08 |
ENST00000370651.3
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr3_-_167813132 | 0.08 |
ENST00000309027.4
|
GOLIM4
|
golgi integral membrane protein 4 |
| chr1_+_53793885 | 0.07 |
ENST00000445039.2
|
RP4-784A16.5
|
RP4-784A16.5 |
| chr6_-_29527702 | 0.07 |
ENST00000377050.4
|
UBD
|
ubiquitin D |
| chr12_-_89920030 | 0.07 |
ENST00000413530.1
ENST00000547474.1 |
GALNT4
POC1B-GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) POC1B-GALNT4 readthrough |
| chr6_+_64282447 | 0.07 |
ENST00000370650.2
ENST00000578299.1 |
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr7_+_123469037 | 0.07 |
ENST00000489978.1
|
HYAL4
|
hyaluronoglucosaminidase 4 |
| chr22_-_42336209 | 0.07 |
ENST00000472374.2
|
CENPM
|
centromere protein M |
| chr10_-_14613968 | 0.07 |
ENST00000488576.1
ENST00000472095.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr17_+_31254892 | 0.07 |
ENST00000394642.3
ENST00000579849.1 |
TMEM98
|
transmembrane protein 98 |
| chr17_-_18266818 | 0.07 |
ENST00000583780.1
|
SHMT1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr10_-_14614311 | 0.07 |
ENST00000479731.1
ENST00000468492.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr15_+_30918879 | 0.07 |
ENST00000428041.2
|
ARHGAP11B
|
Rho GTPase activating protein 11B |
| chr16_+_2198604 | 0.07 |
ENST00000210187.6
|
RAB26
|
RAB26, member RAS oncogene family |
| chr7_+_142829162 | 0.06 |
ENST00000291009.3
|
PIP
|
prolactin-induced protein |
| chr17_+_68101117 | 0.06 |
ENST00000587698.1
ENST00000587892.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chrX_+_38420783 | 0.06 |
ENST00000422612.2
ENST00000286824.6 ENST00000545599.1 |
TSPAN7
|
tetraspanin 7 |
| chr2_+_179317994 | 0.06 |
ENST00000375129.4
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr17_-_63556414 | 0.06 |
ENST00000585045.1
|
AXIN2
|
axin 2 |
| chr2_+_69240302 | 0.06 |
ENST00000303714.4
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr2_+_172378757 | 0.06 |
ENST00000409484.1
ENST00000321348.4 ENST00000375252.3 |
CYBRD1
|
cytochrome b reductase 1 |
| chr14_+_59951161 | 0.06 |
ENST00000261247.9
ENST00000425728.2 ENST00000556985.1 ENST00000554271.1 ENST00000554795.1 |
JKAMP
|
JNK1/MAPK8-associated membrane protein |
| chr15_+_63414760 | 0.06 |
ENST00000557972.1
|
LACTB
|
lactamase, beta |
| chrX_-_106960285 | 0.06 |
ENST00000503515.1
ENST00000372397.2 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr7_-_36406750 | 0.06 |
ENST00000453212.1
ENST00000415803.2 ENST00000440378.1 ENST00000431396.1 ENST00000317020.6 ENST00000436884.1 |
KIAA0895
|
KIAA0895 |
| chr16_-_15736881 | 0.06 |
ENST00000540441.2
|
KIAA0430
|
KIAA0430 |
| chr19_+_41949054 | 0.06 |
ENST00000378187.2
|
C19orf69
|
chromosome 19 open reading frame 69 |
| chr7_+_89975979 | 0.06 |
ENST00000257659.8
ENST00000222511.6 ENST00000417207.1 |
GTPBP10
|
GTP-binding protein 10 (putative) |
| chr5_+_43603229 | 0.06 |
ENST00000344920.4
ENST00000512996.2 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr7_+_151791095 | 0.06 |
ENST00000422997.2
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr12_+_104359576 | 0.06 |
ENST00000392872.3
ENST00000436021.2 |
TDG
|
thymine-DNA glycosylase |
| chr8_+_73921085 | 0.06 |
ENST00000276603.5
ENST00000276602.6 ENST00000518874.1 |
TERF1
|
telomeric repeat binding factor (NIMA-interacting) 1 |
| chr14_-_71107921 | 0.05 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr6_-_112408661 | 0.05 |
ENST00000368662.5
|
TUBE1
|
tubulin, epsilon 1 |
| chr2_+_46926326 | 0.05 |
ENST00000394861.2
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr2_+_24150180 | 0.05 |
ENST00000404924.1
|
UBXN2A
|
UBX domain protein 2A |
| chr4_+_95128996 | 0.05 |
ENST00000457823.2
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr22_+_29168652 | 0.05 |
ENST00000249064.4
ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117
|
coiled-coil domain containing 117 |
| chrX_+_120181457 | 0.05 |
ENST00000328078.1
|
GLUD2
|
glutamate dehydrogenase 2 |
| chr4_+_95972822 | 0.05 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr17_+_31255023 | 0.05 |
ENST00000395149.2
ENST00000261713.4 |
TMEM98
|
transmembrane protein 98 |
| chr19_-_14992264 | 0.05 |
ENST00000327462.2
|
OR7A17
|
olfactory receptor, family 7, subfamily A, member 17 |
| chr16_+_21244986 | 0.05 |
ENST00000311620.5
|
ANKS4B
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr7_+_139528952 | 0.05 |
ENST00000416849.2
ENST00000436047.2 ENST00000414508.2 ENST00000448866.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr2_-_136288740 | 0.05 |
ENST00000264159.6
ENST00000536680.1 |
ZRANB3
|
zinc finger, RAN-binding domain containing 3 |
| chr11_+_100862811 | 0.05 |
ENST00000303130.2
|
TMEM133
|
transmembrane protein 133 |
| chr14_-_21502944 | 0.05 |
ENST00000382951.3
|
RNASE13
|
ribonuclease, RNase A family, 13 (non-active) |
| chr17_+_57697246 | 0.05 |
ENST00000580081.1
ENST00000579456.1 |
CLTC
|
clathrin, heavy chain (Hc) |
| chr14_-_74485960 | 0.05 |
ENST00000556242.1
ENST00000334696.6 |
ENTPD5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr7_+_111846643 | 0.05 |
ENST00000361822.3
|
ZNF277
|
zinc finger protein 277 |
| chr6_+_125540951 | 0.05 |
ENST00000524679.1
|
TPD52L1
|
tumor protein D52-like 1 |
| chr3_-_120068143 | 0.05 |
ENST00000295628.3
|
LRRC58
|
leucine rich repeat containing 58 |
| chr11_-_33774944 | 0.05 |
ENST00000532057.1
ENST00000531080.1 |
FBXO3
|
F-box protein 3 |
| chr8_-_2585929 | 0.05 |
ENST00000519393.1
ENST00000520842.1 ENST00000520570.1 ENST00000517357.1 ENST00000517984.1 ENST00000523971.1 |
RP11-134O21.1
|
RP11-134O21.1 |
| chr20_-_17511962 | 0.04 |
ENST00000377873.3
|
BFSP1
|
beaded filament structural protein 1, filensin |
| chr2_+_177134201 | 0.04 |
ENST00000452865.1
|
MTX2
|
metaxin 2 |
| chr1_+_150229554 | 0.04 |
ENST00000369111.4
|
CA14
|
carbonic anhydrase XIV |
| chr1_+_183774240 | 0.04 |
ENST00000360851.3
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1 |
| chr8_-_101962777 | 0.04 |
ENST00000395951.3
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chrX_+_100663243 | 0.04 |
ENST00000316594.5
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H') |
| chr10_-_116444371 | 0.04 |
ENST00000533213.2
ENST00000369252.4 |
ABLIM1
|
actin binding LIM protein 1 |
| chr12_-_112614506 | 0.04 |
ENST00000548588.2
|
HECTD4
|
HECT domain containing E3 ubiquitin protein ligase 4 |
| chr8_+_38644778 | 0.04 |
ENST00000276520.8
|
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr3_+_36421826 | 0.04 |
ENST00000273183.3
|
STAC
|
SH3 and cysteine rich domain |
| chr16_+_72088376 | 0.04 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr12_-_46385811 | 0.04 |
ENST00000419565.2
|
SCAF11
|
SR-related CTD-associated factor 11 |
| chr13_+_45694583 | 0.04 |
ENST00000340473.6
|
GTF2F2
|
general transcription factor IIF, polypeptide 2, 30kDa |
| chr2_-_86564776 | 0.04 |
ENST00000165698.5
ENST00000541910.1 ENST00000535845.1 |
REEP1
|
receptor accessory protein 1 |
| chr22_+_31742875 | 0.04 |
ENST00000504184.2
|
AC005003.1
|
CDNA FLJ20464 fis, clone KAT06158; HCG1777549; Uncharacterized protein |
| chrX_-_128977364 | 0.04 |
ENST00000371064.3
|
ZDHHC9
|
zinc finger, DHHC-type containing 9 |
| chr17_-_46623441 | 0.04 |
ENST00000330070.4
|
HOXB2
|
homeobox B2 |
| chr15_-_35280426 | 0.04 |
ENST00000559564.1
ENST00000356321.4 |
ZNF770
|
zinc finger protein 770 |
| chr22_-_47134077 | 0.04 |
ENST00000541677.1
ENST00000216264.8 |
CERK
|
ceramide kinase |
| chr6_+_116937636 | 0.04 |
ENST00000368581.4
ENST00000229554.5 ENST00000368580.4 |
RSPH4A
|
radial spoke head 4 homolog A (Chlamydomonas) |
| chr10_-_14614095 | 0.04 |
ENST00000482277.1
ENST00000378462.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr17_+_76037081 | 0.04 |
ENST00000588549.1
|
TNRC6C
|
trinucleotide repeat containing 6C |
| chr22_+_39493268 | 0.04 |
ENST00000401756.1
|
APOBEC3H
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3H |
| chr8_-_17579726 | 0.04 |
ENST00000381861.3
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr10_-_14614122 | 0.04 |
ENST00000378465.3
ENST00000452706.2 ENST00000378458.2 |
FAM107B
|
family with sequence similarity 107, member B |
| chr20_+_5987890 | 0.04 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr4_-_71705027 | 0.04 |
ENST00000545193.1
|
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chr2_+_189157536 | 0.04 |
ENST00000409580.1
ENST00000409637.3 |
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr20_-_656437 | 0.04 |
ENST00000488788.2
|
RP5-850E9.3
|
Uncharacterized protein |
| chr7_+_130131907 | 0.04 |
ENST00000223215.4
ENST00000437945.1 |
MEST
|
mesoderm specific transcript |
| chr4_+_95129061 | 0.04 |
ENST00000354268.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr7_-_148725544 | 0.04 |
ENST00000413966.1
|
PDIA4
|
protein disulfide isomerase family A, member 4 |
| chr4_-_71705060 | 0.04 |
ENST00000514161.1
|
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chr19_+_21579908 | 0.04 |
ENST00000596302.1
ENST00000392288.2 ENST00000594390.1 ENST00000355504.4 |
ZNF493
|
zinc finger protein 493 |
| chr11_-_76155618 | 0.04 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr15_+_62359175 | 0.04 |
ENST00000355522.5
|
C2CD4A
|
C2 calcium-dependent domain containing 4A |
| chr15_+_52043758 | 0.04 |
ENST00000249700.4
ENST00000539962.2 |
TMOD2
|
tropomodulin 2 (neuronal) |
| chr10_-_104211294 | 0.04 |
ENST00000239125.1
|
C10orf95
|
chromosome 10 open reading frame 95 |
| chr9_+_124088860 | 0.04 |
ENST00000373806.1
|
GSN
|
gelsolin |
| chr10_+_104629199 | 0.04 |
ENST00000369880.3
|
AS3MT
|
arsenic (+3 oxidation state) methyltransferase |
| chr2_-_198299726 | 0.03 |
ENST00000409915.4
ENST00000487698.1 ENST00000414963.2 ENST00000335508.6 |
SF3B1
|
splicing factor 3b, subunit 1, 155kDa |
| chr12_+_49717019 | 0.03 |
ENST00000549275.1
ENST00000551245.1 ENST00000380327.5 ENST00000548311.1 ENST00000550346.1 ENST00000550709.1 ENST00000549534.1 ENST00000257909.3 |
TROAP
|
trophinin associated protein |
| chr2_-_29093132 | 0.03 |
ENST00000306108.5
|
TRMT61B
|
tRNA methyltransferase 61 homolog B (S. cerevisiae) |
| chr5_-_79287060 | 0.03 |
ENST00000512560.1
ENST00000509852.1 ENST00000512528.1 |
MTX3
|
metaxin 3 |
| chr2_+_158114051 | 0.03 |
ENST00000259056.4
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr3_+_133502877 | 0.03 |
ENST00000466490.2
|
SRPRB
|
signal recognition particle receptor, B subunit |
| chr22_+_39077947 | 0.03 |
ENST00000216034.4
|
TOMM22
|
translocase of outer mitochondrial membrane 22 homolog (yeast) |
| chr17_+_31255165 | 0.03 |
ENST00000578289.1
ENST00000439138.1 |
TMEM98
|
transmembrane protein 98 |
| chr7_+_112120908 | 0.03 |
ENST00000439068.2
ENST00000312849.4 ENST00000429049.1 |
LSMEM1
|
leucine-rich single-pass membrane protein 1 |
| chr9_+_125795788 | 0.03 |
ENST00000373643.5
|
RABGAP1
|
RAB GTPase activating protein 1 |
| chr15_-_98646940 | 0.03 |
ENST00000560195.1
|
CTD-2544M6.2
|
CTD-2544M6.2 |
| chr1_+_166808667 | 0.03 |
ENST00000537173.1
ENST00000536514.1 ENST00000449930.1 |
POGK
|
pogo transposable element with KRAB domain |
| chr4_-_146019693 | 0.03 |
ENST00000514390.1
|
ANAPC10
|
anaphase promoting complex subunit 10 |
| chr5_-_157286104 | 0.03 |
ENST00000530742.1
ENST00000523908.1 ENST00000523094.1 ENST00000296951.5 ENST00000411809.2 |
CLINT1
|
clathrin interactor 1 |
| chr16_-_46864955 | 0.03 |
ENST00000565112.1
|
C16orf87
|
chromosome 16 open reading frame 87 |
| chr1_+_156254070 | 0.03 |
ENST00000405535.2
ENST00000456810.1 |
TMEM79
|
transmembrane protein 79 |
| chr15_+_52043813 | 0.03 |
ENST00000435126.2
|
TMOD2
|
tropomodulin 2 (neuronal) |
| chr1_-_173991434 | 0.03 |
ENST00000367696.2
|
RC3H1
|
ring finger and CCCH-type domains 1 |
| chr10_+_7745232 | 0.03 |
ENST00000358415.4
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr17_+_75283973 | 0.03 |
ENST00000431235.2
ENST00000449803.2 |
SEPT9
|
septin 9 |
| chr5_+_140165876 | 0.03 |
ENST00000504120.2
ENST00000394633.3 ENST00000378133.3 |
PCDHA1
|
protocadherin alpha 1 |
| chr8_-_119964434 | 0.03 |
ENST00000297350.4
|
TNFRSF11B
|
tumor necrosis factor receptor superfamily, member 11b |
| chr1_-_211752073 | 0.03 |
ENST00000367001.4
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1 |
| chr12_+_27677085 | 0.03 |
ENST00000545334.1
ENST00000540114.1 ENST00000537927.1 ENST00000318304.8 ENST00000535047.1 ENST00000542629.1 ENST00000228425.6 |
PPFIBP1
|
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
| chrX_-_63425561 | 0.03 |
ENST00000374869.3
ENST00000330258.3 |
AMER1
|
APC membrane recruitment protein 1 |
| chr4_-_109541539 | 0.03 |
ENST00000509984.1
ENST00000507248.1 ENST00000506795.1 |
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr11_-_111383064 | 0.03 |
ENST00000525791.1
ENST00000456861.2 ENST00000356018.2 |
BTG4
|
B-cell translocation gene 4 |
| chr2_-_238499337 | 0.03 |
ENST00000411462.1
ENST00000409822.1 |
RAB17
|
RAB17, member RAS oncogene family |
| chr2_-_136288113 | 0.03 |
ENST00000401392.1
|
ZRANB3
|
zinc finger, RAN-binding domain containing 3 |
| chr7_-_135433460 | 0.03 |
ENST00000415751.1
|
FAM180A
|
family with sequence similarity 180, member A |
| chr9_+_103189405 | 0.03 |
ENST00000395067.2
|
MSANTD3
|
Myb/SANT-like DNA-binding domain containing 3 |
| chr12_+_104359641 | 0.03 |
ENST00000537100.1
|
TDG
|
thymine-DNA glycosylase |
| chr6_-_25874440 | 0.03 |
ENST00000361703.6
ENST00000397060.4 |
SLC17A3
|
solute carrier family 17 (organic anion transporter), member 3 |
| chr8_-_12612962 | 0.03 |
ENST00000398246.3
|
LONRF1
|
LON peptidase N-terminal domain and ring finger 1 |
| chr17_-_56065484 | 0.03 |
ENST00000581208.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr1_-_155162658 | 0.03 |
ENST00000368389.2
ENST00000368396.4 ENST00000343256.5 ENST00000342482.4 ENST00000368398.3 ENST00000368390.3 ENST00000337604.5 ENST00000368392.3 ENST00000438413.1 ENST00000368393.3 ENST00000457295.2 ENST00000338684.5 ENST00000368395.1 |
MUC1
|
mucin 1, cell surface associated |
| chr1_-_219615984 | 0.03 |
ENST00000420762.1
|
RP11-95P13.1
|
RP11-95P13.1 |
| chr10_+_7745303 | 0.03 |
ENST00000429820.1
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr15_-_33447055 | 0.03 |
ENST00000559047.1
ENST00000561249.1 |
FMN1
|
formin 1 |
| chr12_+_49717081 | 0.03 |
ENST00000547807.1
ENST00000551567.1 |
TROAP
|
trophinin associated protein |
| chr11_-_129062093 | 0.03 |
ENST00000310343.9
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr12_-_49582593 | 0.03 |
ENST00000295766.5
|
TUBA1A
|
tubulin, alpha 1a |
| chr6_+_108881012 | 0.03 |
ENST00000343882.6
|
FOXO3
|
forkhead box O3 |
| chr6_-_27440460 | 0.03 |
ENST00000377419.1
|
ZNF184
|
zinc finger protein 184 |
| chrX_-_16881144 | 0.03 |
ENST00000416035.1
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr8_-_134511587 | 0.03 |
ENST00000523855.1
ENST00000523854.1 |
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr10_+_17271266 | 0.03 |
ENST00000224237.5
|
VIM
|
vimentin |
| chr13_+_42846272 | 0.03 |
ENST00000025301.2
|
AKAP11
|
A kinase (PRKA) anchor protein 11 |
| chr2_+_234621551 | 0.03 |
ENST00000608381.1
ENST00000373414.3 |
UGT1A1
UGT1A5
|
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr1_+_68150744 | 0.03 |
ENST00000370986.4
ENST00000370985.3 |
GADD45A
|
growth arrest and DNA-damage-inducible, alpha |
| chr6_+_64346386 | 0.03 |
ENST00000509330.1
|
PHF3
|
PHD finger protein 3 |
| chr11_-_8615687 | 0.02 |
ENST00000534493.1
ENST00000422559.2 |
STK33
|
serine/threonine kinase 33 |
| chr9_+_125703282 | 0.02 |
ENST00000373647.4
ENST00000402311.1 |
RABGAP1
|
RAB GTPase activating protein 1 |
| chr4_-_103266219 | 0.02 |
ENST00000394833.2
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr11_-_8615488 | 0.02 |
ENST00000315204.1
ENST00000396672.1 ENST00000396673.1 |
STK33
|
serine/threonine kinase 33 |
| chr1_-_75198940 | 0.02 |
ENST00000417775.1
|
CRYZ
|
crystallin, zeta (quinone reductase) |
| chr6_+_41514305 | 0.02 |
ENST00000409208.1
ENST00000373057.3 |
FOXP4
|
forkhead box P4 |
| chr1_-_75198681 | 0.02 |
ENST00000370872.3
ENST00000370871.3 ENST00000340866.5 ENST00000370870.1 |
CRYZ
|
crystallin, zeta (quinone reductase) |
| chr7_-_87505658 | 0.02 |
ENST00000341119.5
|
SLC25A40
|
solute carrier family 25, member 40 |
| chr15_+_36871806 | 0.02 |
ENST00000566621.1
ENST00000564586.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chrX_-_102757802 | 0.02 |
ENST00000372633.1
|
RAB40A
|
RAB40A, member RAS oncogene family |
| chr11_+_50257784 | 0.02 |
ENST00000529219.1
|
RP11-347H15.4
|
RP11-347H15.4 |
| chr6_-_27440837 | 0.02 |
ENST00000211936.6
|
ZNF184
|
zinc finger protein 184 |
| chr11_+_46740730 | 0.02 |
ENST00000311907.5
ENST00000530231.1 ENST00000442468.1 |
F2
|
coagulation factor II (thrombin) |
| chr7_+_73868439 | 0.02 |
ENST00000424337.2
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr7_+_134576151 | 0.02 |
ENST00000393118.2
|
CALD1
|
caldesmon 1 |
| chr12_-_133405288 | 0.02 |
ENST00000204726.3
|
GOLGA3
|
golgin A3 |
| chr22_-_39052300 | 0.02 |
ENST00000355830.6
|
FAM227A
|
family with sequence similarity 227, member A |
| chr17_+_79679299 | 0.02 |
ENST00000331531.5
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr12_-_133405409 | 0.02 |
ENST00000545875.1
ENST00000456883.2 |
GOLGA3
|
golgin A3 |
| chr2_-_36825281 | 0.02 |
ENST00000405912.3
ENST00000379245.4 |
FEZ2
|
fasciculation and elongation protein zeta 2 (zygin II) |
| chr3_+_169490606 | 0.02 |
ENST00000349841.5
|
MYNN
|
myoneurin |
| chr12_-_11150474 | 0.02 |
ENST00000538986.1
|
TAS2R20
|
taste receptor, type 2, member 20 |
| chr4_-_52904425 | 0.02 |
ENST00000535450.1
|
SGCB
|
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
Network of associatons between targets according to the STRING database.
First level regulatory network of TCF7
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.0 | 0.1 | GO:0090381 | negative regulation of mesodermal cell fate specification(GO:0042662) regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) |
| 0.0 | 0.1 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.1 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.1 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.0 | 0.1 | GO:1904482 | response to tetrahydrofolate(GO:1904481) cellular response to tetrahydrofolate(GO:1904482) |
| 0.0 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.1 | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.1 | GO:1903899 | lung goblet cell differentiation(GO:0060480) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.0 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.0 | 0.0 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.0 | 0.0 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.0 | 0.0 | GO:0071423 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.0 | 0.1 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.0 | 0.0 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.0 | GO:0001534 | radial spoke(GO:0001534) |
| 0.0 | 0.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) |
| 0.0 | 0.0 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.0 | 0.0 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.0 | GO:0043337 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 0.0 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.1 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.0 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |