Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
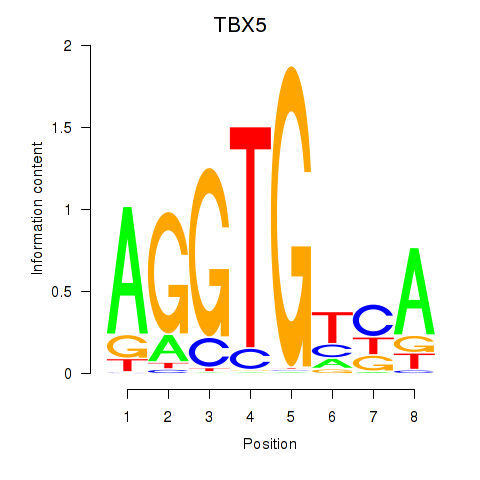
Results for TBX5
Z-value: 1.17
Transcription factors associated with TBX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX5
|
ENSG00000089225.15 | T-box transcription factor 5 |
Activity profile of TBX5 motif
Sorted Z-values of TBX5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_39786962 | 2.20 |
ENST00000333625.2
|
IFNL1
|
interferon, lambda 1 |
| chr14_-_37051798 | 1.52 |
ENST00000258829.5
|
NKX2-8
|
NK2 homeobox 8 |
| chr17_+_68165657 | 1.35 |
ENST00000243457.3
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr16_-_11350036 | 1.27 |
ENST00000332029.2
|
SOCS1
|
suppressor of cytokine signaling 1 |
| chr8_-_145060593 | 1.15 |
ENST00000313059.5
ENST00000524918.1 ENST00000313028.7 ENST00000525773.1 |
PARP10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr2_-_191878874 | 1.01 |
ENST00000392322.3
ENST00000392323.2 ENST00000424722.1 ENST00000361099.3 |
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr11_+_117947724 | 0.99 |
ENST00000534111.1
|
TMPRSS4
|
transmembrane protease, serine 4 |
| chr2_-_42160486 | 0.95 |
ENST00000427054.1
|
AC104654.2
|
AC104654.2 |
| chr5_-_134914673 | 0.89 |
ENST00000512158.1
|
CXCL14
|
chemokine (C-X-C motif) ligand 14 |
| chr15_+_89182156 | 0.85 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr3_-_182880541 | 0.83 |
ENST00000470251.1
ENST00000265598.3 |
LAMP3
|
lysosomal-associated membrane protein 3 |
| chr15_+_89182178 | 0.80 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr16_+_57023406 | 0.76 |
ENST00000262510.6
ENST00000308149.7 ENST00000436936.1 |
NLRC5
|
NLR family, CARD domain containing 5 |
| chr1_+_15736359 | 0.74 |
ENST00000375980.4
|
EFHD2
|
EF-hand domain family, member D2 |
| chr20_-_4055812 | 0.72 |
ENST00000379526.1
|
RP11-352D3.2
|
Uncharacterized protein |
| chr22_-_20231207 | 0.72 |
ENST00000425986.1
|
RTN4R
|
reticulon 4 receptor |
| chr15_+_89181974 | 0.62 |
ENST00000306072.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr6_+_83073952 | 0.60 |
ENST00000543496.1
|
TPBG
|
trophoblast glycoprotein |
| chr19_-_10213335 | 0.60 |
ENST00000592641.1
ENST00000253109.4 |
ANGPTL6
|
angiopoietin-like 6 |
| chr6_-_26027480 | 0.59 |
ENST00000377364.3
|
HIST1H4B
|
histone cluster 1, H4b |
| chr22_+_38597889 | 0.58 |
ENST00000338483.2
ENST00000538320.1 ENST00000538999.1 ENST00000441709.1 |
MAFF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr11_-_115127611 | 0.57 |
ENST00000545094.1
|
CADM1
|
cell adhesion molecule 1 |
| chr17_+_74372662 | 0.56 |
ENST00000591651.1
ENST00000545180.1 |
SPHK1
|
sphingosine kinase 1 |
| chr3_-_120170052 | 0.56 |
ENST00000295633.3
|
FSTL1
|
follistatin-like 1 |
| chr1_+_8378140 | 0.51 |
ENST00000377479.2
|
SLC45A1
|
solute carrier family 45, member 1 |
| chr2_+_101591314 | 0.50 |
ENST00000450763.1
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr2_+_5832799 | 0.50 |
ENST00000322002.3
|
SOX11
|
SRY (sex determining region Y)-box 11 |
| chr22_-_51016433 | 0.48 |
ENST00000405237.3
|
CPT1B
|
carnitine palmitoyltransferase 1B (muscle) |
| chr11_-_45928830 | 0.48 |
ENST00000449465.1
|
C11orf94
|
chromosome 11 open reading frame 94 |
| chr19_+_18723660 | 0.46 |
ENST00000262817.3
|
TMEM59L
|
transmembrane protein 59-like |
| chr8_-_123793048 | 0.44 |
ENST00000607710.1
|
RP11-44N11.2
|
RP11-44N11.2 |
| chr22_-_51016846 | 0.44 |
ENST00000312108.7
ENST00000395650.2 |
CPT1B
|
carnitine palmitoyltransferase 1B (muscle) |
| chr12_-_116714564 | 0.43 |
ENST00000548743.1
|
MED13L
|
mediator complex subunit 13-like |
| chr6_-_19804973 | 0.43 |
ENST00000457670.1
ENST00000607810.1 ENST00000606628.1 |
RP4-625H18.2
|
RP4-625H18.2 |
| chr8_+_32405728 | 0.42 |
ENST00000523079.1
ENST00000338921.4 ENST00000356819.4 ENST00000287845.5 ENST00000341377.5 |
NRG1
|
neuregulin 1 |
| chr5_-_74348371 | 0.42 |
ENST00000503568.1
|
RP11-229C3.2
|
RP11-229C3.2 |
| chr9_-_96717654 | 0.42 |
ENST00000253968.6
|
BARX1
|
BARX homeobox 1 |
| chr17_-_79817091 | 0.42 |
ENST00000570907.1
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr1_-_12677714 | 0.41 |
ENST00000376223.2
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr22_+_31488433 | 0.40 |
ENST00000455608.1
|
SMTN
|
smoothelin |
| chr11_-_74660065 | 0.39 |
ENST00000525407.1
ENST00000528219.1 ENST00000531852.1 |
XRRA1
|
X-ray radiation resistance associated 1 |
| chr1_-_153521714 | 0.38 |
ENST00000368713.3
|
S100A3
|
S100 calcium binding protein A3 |
| chr16_-_67190152 | 0.37 |
ENST00000486556.1
|
TRADD
|
TNFRSF1A-associated via death domain |
| chr8_+_32405785 | 0.37 |
ENST00000287842.3
|
NRG1
|
neuregulin 1 |
| chr19_+_41107249 | 0.36 |
ENST00000396819.3
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr5_-_180229791 | 0.36 |
ENST00000504671.1
ENST00000507384.1 |
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr11_-_134095335 | 0.35 |
ENST00000534227.1
ENST00000532445.1 |
NCAPD3
|
non-SMC condensin II complex, subunit D3 |
| chr3_+_183899770 | 0.35 |
ENST00000442686.1
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit |
| chr12_+_112563303 | 0.35 |
ENST00000412615.2
|
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chr17_-_73937028 | 0.34 |
ENST00000586631.2
|
FBF1
|
Fas (TNFRSF6) binding factor 1 |
| chr4_-_140222358 | 0.34 |
ENST00000505036.1
ENST00000544855.1 ENST00000539002.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr12_+_19358192 | 0.34 |
ENST00000538305.1
|
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr11_-_125932685 | 0.34 |
ENST00000527967.1
|
CDON
|
cell adhesion associated, oncogene regulated |
| chr18_+_33877654 | 0.33 |
ENST00000257209.4
ENST00000445677.1 ENST00000590592.1 ENST00000359247.4 |
FHOD3
|
formin homology 2 domain containing 3 |
| chr17_-_26220366 | 0.33 |
ENST00000460380.2
ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9
RP1-66C13.4
|
LYR motif containing 9 Uncharacterized protein |
| chr17_+_77020146 | 0.33 |
ENST00000579760.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr17_+_40119801 | 0.33 |
ENST00000585452.1
|
CNP
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr11_+_58390132 | 0.33 |
ENST00000361987.4
|
CNTF
|
ciliary neurotrophic factor |
| chr6_+_122931366 | 0.32 |
ENST00000368452.2
ENST00000368448.1 ENST00000392490.1 |
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr15_+_41099254 | 0.32 |
ENST00000570108.1
ENST00000564258.1 ENST00000355341.4 ENST00000336455.5 |
ZFYVE19
|
zinc finger, FYVE domain containing 19 |
| chr3_-_48632593 | 0.32 |
ENST00000454817.1
ENST00000328333.8 |
COL7A1
|
collagen, type VII, alpha 1 |
| chr12_-_49351303 | 0.31 |
ENST00000256682.4
|
ARF3
|
ADP-ribosylation factor 3 |
| chr17_+_77020325 | 0.31 |
ENST00000311661.4
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr17_+_77020224 | 0.31 |
ENST00000339142.2
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr17_-_72968837 | 0.31 |
ENST00000581676.1
|
HID1
|
HID1 domain containing |
| chr22_+_38609538 | 0.31 |
ENST00000407965.1
|
MAFF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr1_+_228395755 | 0.31 |
ENST00000284548.11
ENST00000570156.2 ENST00000422127.1 ENST00000366707.4 ENST00000366709.4 |
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr4_+_81118647 | 0.31 |
ENST00000415738.2
|
PRDM8
|
PR domain containing 8 |
| chr20_+_45523227 | 0.30 |
ENST00000327619.5
ENST00000357410.3 |
EYA2
|
eyes absent homolog 2 (Drosophila) |
| chr17_+_57970469 | 0.30 |
ENST00000443572.2
ENST00000406116.3 ENST00000225577.4 ENST00000393021.3 |
RPS6KB1
|
ribosomal protein S6 kinase, 70kDa, polypeptide 1 |
| chr17_-_15469590 | 0.30 |
ENST00000312127.2
|
CDRT1
|
CMT duplicated region transcript 1; Uncharacterized protein |
| chr11_+_46402297 | 0.30 |
ENST00000405308.2
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chrX_-_19689106 | 0.29 |
ENST00000379716.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr15_+_91416092 | 0.29 |
ENST00000559353.1
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chr2_-_61697862 | 0.29 |
ENST00000398571.2
|
USP34
|
ubiquitin specific peptidase 34 |
| chr12_+_112563335 | 0.29 |
ENST00000549358.1
ENST00000257604.5 ENST00000548092.1 ENST00000552896.1 |
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chr16_+_57139933 | 0.29 |
ENST00000566259.1
|
CPNE2
|
copine II |
| chr11_+_120081475 | 0.29 |
ENST00000328965.4
|
OAF
|
OAF homolog (Drosophila) |
| chr10_+_135340859 | 0.29 |
ENST00000252945.3
ENST00000421586.1 ENST00000418356.1 |
CYP2E1
|
cytochrome P450, family 2, subfamily E, polypeptide 1 |
| chr19_+_1000418 | 0.28 |
ENST00000234389.3
|
GRIN3B
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3B |
| chr17_+_79859985 | 0.28 |
ENST00000333383.7
|
NPB
|
neuropeptide B |
| chrX_-_83757399 | 0.27 |
ENST00000373177.2
ENST00000297977.5 ENST00000506585.2 ENST00000449553.2 |
HDX
|
highly divergent homeobox |
| chr20_-_45530365 | 0.27 |
ENST00000414085.1
|
RP11-323C15.2
|
RP11-323C15.2 |
| chr10_-_125853200 | 0.26 |
ENST00000421115.1
|
CHST15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
| chr19_+_54385439 | 0.26 |
ENST00000536044.1
ENST00000540413.1 ENST00000263431.3 ENST00000419486.1 |
PRKCG
|
protein kinase C, gamma |
| chr1_-_149982624 | 0.26 |
ENST00000417191.1
ENST00000369135.4 |
OTUD7B
|
OTU domain containing 7B |
| chr19_+_10397648 | 0.26 |
ENST00000340992.4
ENST00000393717.2 |
ICAM4
|
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
| chr22_-_51017084 | 0.26 |
ENST00000360719.2
ENST00000457250.1 ENST00000440709.1 |
CPT1B
|
carnitine palmitoyltransferase 1B (muscle) |
| chr17_+_77021702 | 0.26 |
ENST00000392445.2
ENST00000354124.3 |
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr7_+_96634850 | 0.26 |
ENST00000518156.2
|
DLX6
|
distal-less homeobox 6 |
| chr21_+_30503282 | 0.26 |
ENST00000399925.1
|
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr4_+_183065793 | 0.26 |
ENST00000512480.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr9_+_96717821 | 0.25 |
ENST00000454594.1
|
RP11-231K24.2
|
RP11-231K24.2 |
| chr3_+_184016986 | 0.25 |
ENST00000417952.1
|
PSMD2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| chr3_-_52719912 | 0.25 |
ENST00000420148.1
|
PBRM1
|
polybromo 1 |
| chr22_-_30722912 | 0.25 |
ENST00000215790.7
|
TBC1D10A
|
TBC1 domain family, member 10A |
| chr19_+_50380917 | 0.25 |
ENST00000535102.2
|
TBC1D17
|
TBC1 domain family, member 17 |
| chr3_-_48594248 | 0.24 |
ENST00000545984.1
ENST00000232375.3 ENST00000416568.1 ENST00000383734.2 ENST00000541519.1 ENST00000412035.1 |
PFKFB4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr20_-_30310336 | 0.24 |
ENST00000434194.1
ENST00000376062.2 |
BCL2L1
|
BCL2-like 1 |
| chr15_+_41913690 | 0.24 |
ENST00000563576.1
|
MGA
|
MGA, MAX dimerization protein |
| chrX_+_102862834 | 0.24 |
ENST00000372627.5
ENST00000243286.3 |
TCEAL3
|
transcription elongation factor A (SII)-like 3 |
| chr6_-_30043539 | 0.23 |
ENST00000376751.3
ENST00000244360.6 |
RNF39
|
ring finger protein 39 |
| chr2_+_171673417 | 0.23 |
ENST00000344257.5
|
GAD1
|
glutamate decarboxylase 1 (brain, 67kDa) |
| chr12_+_57854274 | 0.23 |
ENST00000528432.1
|
GLI1
|
GLI family zinc finger 1 |
| chr17_-_7297833 | 0.23 |
ENST00000571802.1
ENST00000576201.1 ENST00000573213.1 ENST00000324822.11 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr11_-_34535297 | 0.23 |
ENST00000532417.1
|
ELF5
|
E74-like factor 5 (ets domain transcription factor) |
| chr1_+_168250194 | 0.23 |
ENST00000367821.3
|
TBX19
|
T-box 19 |
| chr6_-_31620095 | 0.23 |
ENST00000424176.1
ENST00000456622.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr2_+_234668894 | 0.23 |
ENST00000305208.5
ENST00000608383.1 ENST00000360418.3 |
UGT1A8
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr16_+_30007524 | 0.23 |
ENST00000567254.1
ENST00000567705.1 |
INO80E
|
INO80 complex subunit E |
| chr16_+_20911174 | 0.22 |
ENST00000568663.1
|
LYRM1
|
LYR motif containing 1 |
| chr15_-_43863695 | 0.22 |
ENST00000439195.1
|
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr2_+_171673072 | 0.22 |
ENST00000358196.3
ENST00000375272.1 |
GAD1
|
glutamate decarboxylase 1 (brain, 67kDa) |
| chr19_-_8579030 | 0.22 |
ENST00000255616.8
ENST00000393927.4 |
ZNF414
|
zinc finger protein 414 |
| chr1_-_47655686 | 0.22 |
ENST00000294338.2
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr1_-_153917700 | 0.22 |
ENST00000368646.2
|
DENND4B
|
DENN/MADD domain containing 4B |
| chr17_-_46507567 | 0.22 |
ENST00000584924.1
|
SKAP1
|
src kinase associated phosphoprotein 1 |
| chr12_-_49351228 | 0.22 |
ENST00000541959.1
ENST00000447318.2 |
ARF3
|
ADP-ribosylation factor 3 |
| chr6_-_31619742 | 0.21 |
ENST00000433828.1
ENST00000456286.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr19_+_50380682 | 0.21 |
ENST00000221543.5
|
TBC1D17
|
TBC1 domain family, member 17 |
| chr1_-_33502528 | 0.21 |
ENST00000354858.6
|
AK2
|
adenylate kinase 2 |
| chr7_-_27135591 | 0.21 |
ENST00000343060.4
ENST00000355633.5 |
HOXA1
|
homeobox A1 |
| chr1_-_205290865 | 0.21 |
ENST00000367157.3
|
NUAK2
|
NUAK family, SNF1-like kinase, 2 |
| chrX_-_80377162 | 0.21 |
ENST00000430960.1
ENST00000447319.1 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chrX_+_56259316 | 0.21 |
ENST00000468660.1
|
KLF8
|
Kruppel-like factor 8 |
| chr1_-_44482979 | 0.21 |
ENST00000360584.2
ENST00000357730.2 ENST00000528803.1 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr16_-_3086927 | 0.20 |
ENST00000572449.1
|
CCDC64B
|
coiled-coil domain containing 64B |
| chr12_+_8234807 | 0.20 |
ENST00000339754.5
|
NECAP1
|
NECAP endocytosis associated 1 |
| chr19_+_55987998 | 0.20 |
ENST00000591164.1
|
ZNF628
|
zinc finger protein 628 |
| chr22_-_30722866 | 0.20 |
ENST00000403477.3
|
TBC1D10A
|
TBC1 domain family, member 10A |
| chr1_+_26737253 | 0.20 |
ENST00000326279.6
|
LIN28A
|
lin-28 homolog A (C. elegans) |
| chr16_+_2546033 | 0.20 |
ENST00000564543.1
ENST00000434757.2 |
RP11-20I23.1
TBC1D24
|
Uncharacterized protein TBC1 domain family, member 24 |
| chr15_-_74043816 | 0.20 |
ENST00000379822.4
|
C15orf59
|
chromosome 15 open reading frame 59 |
| chr11_-_119252425 | 0.20 |
ENST00000260187.2
|
USP2
|
ubiquitin specific peptidase 2 |
| chr6_-_31619697 | 0.20 |
ENST00000434444.1
|
BAG6
|
BCL2-associated athanogene 6 |
| chr9_-_16276311 | 0.20 |
ENST00000380685.1
|
C9orf92
|
chromosome 9 open reading frame 92 |
| chr7_-_105319536 | 0.20 |
ENST00000477775.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr1_+_41447609 | 0.20 |
ENST00000543104.1
|
CTPS1
|
CTP synthase 1 |
| chr6_-_19804877 | 0.20 |
ENST00000447250.1
|
RP4-625H18.2
|
RP4-625H18.2 |
| chr12_-_53228079 | 0.20 |
ENST00000330553.5
|
KRT79
|
keratin 79 |
| chr17_+_16945820 | 0.20 |
ENST00000577514.1
|
MPRIP
|
myosin phosphatase Rho interacting protein |
| chr2_-_218766698 | 0.19 |
ENST00000439083.1
|
TNS1
|
tensin 1 |
| chr19_+_10397621 | 0.19 |
ENST00000380770.3
|
ICAM4
|
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
| chrX_+_33744503 | 0.19 |
ENST00000439992.1
|
RP11-305F18.1
|
RP11-305F18.1 |
| chr16_+_2588012 | 0.19 |
ENST00000354836.5
ENST00000389224.3 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr11_-_72385437 | 0.19 |
ENST00000418754.2
ENST00000542969.2 ENST00000334456.5 |
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated |
| chr19_-_41942344 | 0.19 |
ENST00000594660.1
|
ATP5SL
|
ATP5S-like |
| chrX_-_148713365 | 0.19 |
ENST00000511776.1
ENST00000507237.1 |
TMEM185A
|
transmembrane protein 185A |
| chr11_+_117947782 | 0.19 |
ENST00000522307.1
ENST00000523251.1 ENST00000437212.3 ENST00000522824.1 ENST00000522151.1 |
TMPRSS4
|
transmembrane protease, serine 4 |
| chr1_-_33502441 | 0.19 |
ENST00000548033.1
ENST00000487289.1 ENST00000373449.2 ENST00000480134.1 ENST00000467905.1 |
AK2
|
adenylate kinase 2 |
| chr19_-_11456935 | 0.19 |
ENST00000590788.1
ENST00000586590.1 ENST00000589555.1 ENST00000586956.1 ENST00000593256.2 ENST00000447337.1 ENST00000591677.1 ENST00000586701.1 ENST00000589655.1 |
TMEM205
RAB3D
|
transmembrane protein 205 RAB3D, member RAS oncogene family |
| chr18_+_3449695 | 0.19 |
ENST00000343820.5
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr1_+_153963227 | 0.18 |
ENST00000368567.4
ENST00000392558.4 |
RPS27
|
ribosomal protein S27 |
| chr5_+_125758813 | 0.18 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr22_-_30662828 | 0.18 |
ENST00000403463.1
ENST00000215781.2 |
OSM
|
oncostatin M |
| chr11_-_72145426 | 0.18 |
ENST00000535990.1
ENST00000437826.2 ENST00000340729.5 |
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr16_+_57844549 | 0.18 |
ENST00000564282.1
|
CTD-2600O9.1
|
uncharacterized protein LOC388282 |
| chr1_+_26737292 | 0.18 |
ENST00000254231.4
|
LIN28A
|
lin-28 homolog A (C. elegans) |
| chr2_+_153191706 | 0.18 |
ENST00000288670.9
|
FMNL2
|
formin-like 2 |
| chr2_+_201994208 | 0.18 |
ENST00000440180.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr3_+_107602030 | 0.18 |
ENST00000494231.1
|
LINC00636
|
long intergenic non-protein coding RNA 636 |
| chr2_-_200322723 | 0.18 |
ENST00000417098.1
|
SATB2
|
SATB homeobox 2 |
| chr5_+_125758865 | 0.18 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr2_+_210636697 | 0.18 |
ENST00000439458.1
ENST00000272845.6 |
UNC80
|
unc-80 homolog (C. elegans) |
| chr1_+_101702417 | 0.18 |
ENST00000305352.6
|
S1PR1
|
sphingosine-1-phosphate receptor 1 |
| chr9_-_79307096 | 0.17 |
ENST00000376717.2
ENST00000223609.6 ENST00000443509.2 |
PRUNE2
|
prune homolog 2 (Drosophila) |
| chr11_-_34535332 | 0.17 |
ENST00000257832.2
ENST00000429939.2 |
ELF5
|
E74-like factor 5 (ets domain transcription factor) |
| chr18_+_54318893 | 0.17 |
ENST00000593058.1
|
WDR7
|
WD repeat domain 7 |
| chr11_-_72145641 | 0.17 |
ENST00000538039.1
ENST00000445069.2 |
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr11_+_114310164 | 0.17 |
ENST00000544196.1
ENST00000539754.1 ENST00000539275.1 |
REXO2
|
RNA exonuclease 2 |
| chr19_-_11456872 | 0.17 |
ENST00000586218.1
|
TMEM205
|
transmembrane protein 205 |
| chr11_+_114310102 | 0.17 |
ENST00000265881.5
|
REXO2
|
RNA exonuclease 2 |
| chr6_-_31620149 | 0.16 |
ENST00000435080.1
ENST00000375976.4 ENST00000441054.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr1_-_151119087 | 0.16 |
ENST00000341697.3
ENST00000368914.3 |
SEMA6C
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chr5_-_142784888 | 0.16 |
ENST00000514699.1
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr16_-_31147020 | 0.16 |
ENST00000568261.1
ENST00000567797.1 ENST00000317508.6 |
PRSS8
|
protease, serine, 8 |
| chr8_+_81397876 | 0.16 |
ENST00000430430.1
|
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr1_+_202995611 | 0.15 |
ENST00000367240.2
|
PPFIA4
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 |
| chr17_+_34431212 | 0.15 |
ENST00000394495.1
|
CCL4
|
chemokine (C-C motif) ligand 4 |
| chr7_-_130080681 | 0.15 |
ENST00000469826.1
|
CEP41
|
centrosomal protein 41kDa |
| chr11_-_119252359 | 0.15 |
ENST00000455332.2
|
USP2
|
ubiquitin specific peptidase 2 |
| chr19_-_11456905 | 0.15 |
ENST00000588560.1
ENST00000592952.1 |
TMEM205
|
transmembrane protein 205 |
| chr22_-_46373004 | 0.15 |
ENST00000339464.4
|
WNT7B
|
wingless-type MMTV integration site family, member 7B |
| chr6_-_31619892 | 0.15 |
ENST00000454165.1
ENST00000428326.1 ENST00000452994.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr17_-_46703826 | 0.15 |
ENST00000550387.1
ENST00000311177.5 |
HOXB9
|
homeobox B9 |
| chr12_-_62586543 | 0.15 |
ENST00000416284.3
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chrX_+_35937843 | 0.15 |
ENST00000297866.5
|
CXorf22
|
chromosome X open reading frame 22 |
| chr6_+_34204642 | 0.15 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr16_-_4465886 | 0.15 |
ENST00000539968.1
|
CORO7
|
coronin 7 |
| chr6_+_43445261 | 0.14 |
ENST00000372444.2
ENST00000372445.5 ENST00000436109.2 ENST00000372454.2 ENST00000442878.2 ENST00000259751.1 ENST00000372452.1 ENST00000372449.1 |
TJAP1
|
tight junction associated protein 1 (peripheral) |
| chr2_+_149402553 | 0.14 |
ENST00000258484.6
ENST00000409654.1 |
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr13_-_79177673 | 0.14 |
ENST00000377208.5
|
POU4F1
|
POU class 4 homeobox 1 |
| chr1_+_13910194 | 0.14 |
ENST00000376057.4
ENST00000510906.1 |
PDPN
|
podoplanin |
| chr8_-_95907423 | 0.14 |
ENST00000396133.3
ENST00000308108.4 |
CCNE2
|
cyclin E2 |
| chr12_-_52887034 | 0.14 |
ENST00000330722.6
|
KRT6A
|
keratin 6A |
| chr22_-_31885514 | 0.14 |
ENST00000397525.1
|
EIF4ENIF1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr4_-_186456766 | 0.14 |
ENST00000284771.6
|
PDLIM3
|
PDZ and LIM domain 3 |
| chr16_+_30662360 | 0.14 |
ENST00000542965.2
|
PRR14
|
proline rich 14 |
| chr19_-_50380536 | 0.14 |
ENST00000391832.3
ENST00000391834.2 ENST00000344175.5 |
AKT1S1
|
AKT1 substrate 1 (proline-rich) |
| chr11_-_72145669 | 0.14 |
ENST00000543042.1
ENST00000294053.3 |
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.2 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.5 | 2.3 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.4 | 1.5 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.3 | 1.0 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.2 | 1.2 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.2 | 0.6 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.2 | 0.5 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.2 | 1.3 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) |
| 0.2 | 1.2 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 1.0 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 0.3 | GO:0046364 | hexose biosynthetic process(GO:0019319) monosaccharide biosynthetic process(GO:0046364) |
| 0.1 | 0.6 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 0.5 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.1 | 0.8 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 0.3 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 | 0.3 | GO:0032904 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.1 | 1.0 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.1 | 0.3 | GO:1900073 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 0.1 | 0.8 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.1 | 0.4 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.1 | 0.3 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.1 | 0.4 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.1 | 0.1 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.1 | 0.2 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 0.2 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 0.3 | GO:0010193 | response to ozone(GO:0010193) |
| 0.1 | 0.4 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 0.3 | GO:0032425 | positive regulation of mismatch repair(GO:0032425) |
| 0.0 | 0.3 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.4 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 1.1 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.1 | GO:0001897 | cytolysis by symbiont of host cells(GO:0001897) |
| 0.0 | 0.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.3 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.1 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.0 | 0.2 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.4 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.3 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.4 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.0 | 0.2 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.1 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.3 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.4 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.2 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.7 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.2 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) inner medullary collecting duct development(GO:0072061) |
| 0.0 | 0.3 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.2 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.1 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.4 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.1 | GO:2000987 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.2 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.1 | GO:0090402 | senescence-associated heterochromatin focus assembly(GO:0035986) oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 0.1 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 0.0 | 0.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.2 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.0 | 0.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.8 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.3 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.5 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.6 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.4 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.0 | 0.1 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.0 | 0.1 | GO:0042377 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.0 | 0.1 | GO:0046778 | modification by virus of host mRNA processing(GO:0046778) |
| 0.0 | 0.2 | GO:0007507 | heart development(GO:0007507) |
| 0.0 | 0.4 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.1 | GO:0002857 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.2 | GO:0060838 | lymphatic endothelial cell fate commitment(GO:0060838) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.1 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.1 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.0 | 0.1 | GO:0061053 | somite development(GO:0061053) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.1 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.1 | GO:0019075 | virus maturation(GO:0019075) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.0 | GO:0021586 | pons maturation(GO:0021586) |
| 0.0 | 0.1 | GO:0072564 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.1 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.0 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.0 | GO:0071288 | carbon dioxide transmembrane transport(GO:0035378) positive regulation of saliva secretion(GO:0046878) cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:1902954 | regulation of early endosome to recycling endosome transport(GO:1902954) |
| 0.0 | 0.1 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.3 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.0 | 0.1 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.2 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.4 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.1 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:0044126 | regulation of growth of symbiont in host(GO:0044126) |
| 0.0 | 0.1 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.6 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.0 | 0.1 | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.0 | 0.1 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.2 | 0.7 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.1 | 0.4 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 0.3 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.1 | 1.0 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 0.3 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 0.3 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.1 | 0.4 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.1 | 0.3 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.1 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.7 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 2.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) symmetric synapse(GO:0032280) |
| 0.0 | 0.3 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.7 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 1.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.3 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.3 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.2 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.0 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.3 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0042025 | host cell nucleus(GO:0042025) |
| 0.0 | 0.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.2 | 0.7 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.1 | 1.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.1 | 1.5 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.1 | 0.3 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.1 | 0.6 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.5 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.2 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.8 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.3 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.1 | 0.3 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.4 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.4 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 1.0 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.4 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 1.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.6 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.1 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.2 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 1.0 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.4 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.2 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 1.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 1.1 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.4 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.2 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 1.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.8 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.3 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.0 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.0 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.2 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.4 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 2.3 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.3 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 1.2 | GO:0005518 | collagen binding(GO:0005518) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.8 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 2.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.8 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.2 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 1.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 2.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.1 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |