Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
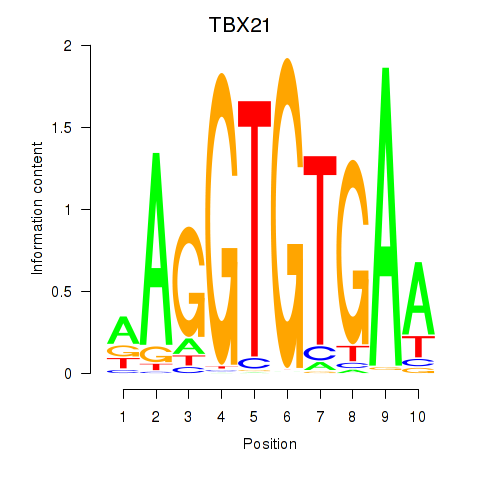
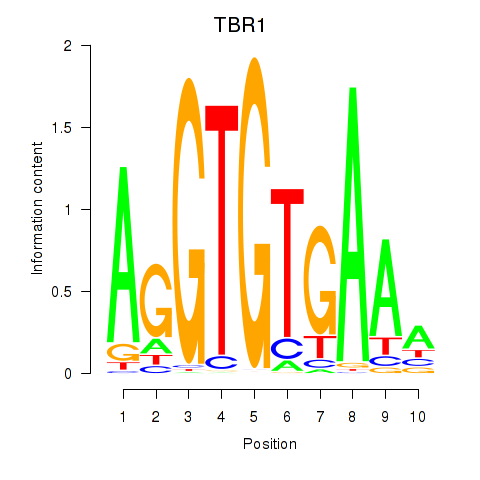
Results for TBX21_TBR1
Z-value: 0.98
Transcription factors associated with TBX21_TBR1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX21
|
ENSG00000073861.2 | T-box transcription factor 21 |
|
TBR1
|
ENSG00000136535.10 | T-box brain transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX21 | hg19_v2_chr17_+_45810594_45810610 | 0.80 | 2.0e-01 | Click! |
| TBR1 | hg19_v2_chr2_+_162272605_162272753 | 0.18 | 8.2e-01 | Click! |
Activity profile of TBX21_TBR1 motif
Sorted Z-values of TBX21_TBR1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_69139467 | 1.48 |
ENST00000569188.1
|
HAS3
|
hyaluronan synthase 3 |
| chr17_-_1553346 | 0.79 |
ENST00000301336.6
|
RILP
|
Rab interacting lysosomal protein |
| chrX_-_106146547 | 0.77 |
ENST00000276173.4
ENST00000411805.1 |
RIPPLY1
|
ripply transcriptional repressor 1 |
| chr19_-_10213335 | 0.73 |
ENST00000592641.1
ENST00000253109.4 |
ANGPTL6
|
angiopoietin-like 6 |
| chr6_-_160166218 | 0.72 |
ENST00000537657.1
|
SOD2
|
superoxide dismutase 2, mitochondrial |
| chr9_+_96717821 | 0.68 |
ENST00000454594.1
|
RP11-231K24.2
|
RP11-231K24.2 |
| chr2_+_176972000 | 0.63 |
ENST00000249504.5
|
HOXD11
|
homeobox D11 |
| chr16_-_11350036 | 0.60 |
ENST00000332029.2
|
SOCS1
|
suppressor of cytokine signaling 1 |
| chr1_+_19967014 | 0.58 |
ENST00000428975.1
|
NBL1
|
neuroblastoma 1, DAN family BMP antagonist |
| chr2_+_228736335 | 0.58 |
ENST00000440997.1
ENST00000545118.1 |
DAW1
|
dynein assembly factor with WDR repeat domains 1 |
| chr9_-_96717654 | 0.57 |
ENST00000253968.6
|
BARX1
|
BARX homeobox 1 |
| chr9_+_127023704 | 0.54 |
ENST00000373596.1
ENST00000425237.1 |
NEK6
|
NIMA-related kinase 6 |
| chr4_+_108746282 | 0.54 |
ENST00000503862.1
|
SGMS2
|
sphingomyelin synthase 2 |
| chr11_-_133826852 | 0.54 |
ENST00000533871.2
ENST00000321016.8 |
IGSF9B
|
immunoglobulin superfamily, member 9B |
| chr22_-_31688431 | 0.54 |
ENST00000402249.3
ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr1_-_44482979 | 0.52 |
ENST00000360584.2
ENST00000357730.2 ENST00000528803.1 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr11_-_615570 | 0.50 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chr5_+_74807886 | 0.49 |
ENST00000514296.1
|
POLK
|
polymerase (DNA directed) kappa |
| chr11_-_33913708 | 0.48 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr1_+_155099927 | 0.46 |
ENST00000368407.3
|
EFNA1
|
ephrin-A1 |
| chr12_+_75874984 | 0.45 |
ENST00000550491.1
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr1_+_39456895 | 0.44 |
ENST00000432648.3
ENST00000446189.2 ENST00000372984.4 |
AKIRIN1
|
akirin 1 |
| chr19_+_10381769 | 0.42 |
ENST00000423829.2
ENST00000588645.1 |
ICAM1
|
intercellular adhesion molecule 1 |
| chr3_+_137728842 | 0.42 |
ENST00000183605.5
|
CLDN18
|
claudin 18 |
| chr14_+_95078714 | 0.42 |
ENST00000393078.3
ENST00000393080.4 ENST00000467132.1 |
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr1_-_100643765 | 0.42 |
ENST00000370137.1
ENST00000370138.1 ENST00000342895.3 |
LRRC39
|
leucine rich repeat containing 39 |
| chr9_+_34329493 | 0.42 |
ENST00000379158.2
ENST00000346365.4 ENST00000379155.5 |
NUDT2
|
nudix (nucleoside diphosphate linked moiety X)-type motif 2 |
| chr14_-_98444386 | 0.42 |
ENST00000556462.1
ENST00000556138.1 |
C14orf64
|
chromosome 14 open reading frame 64 |
| chrX_+_56259316 | 0.41 |
ENST00000468660.1
|
KLF8
|
Kruppel-like factor 8 |
| chr5_-_39270725 | 0.41 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr16_+_66461175 | 0.40 |
ENST00000536005.2
ENST00000299694.8 ENST00000561796.1 |
BEAN1
|
brain expressed, associated with NEDD4, 1 |
| chr2_-_152146385 | 0.40 |
ENST00000414946.1
ENST00000243346.5 |
NMI
|
N-myc (and STAT) interactor |
| chr1_-_205290865 | 0.40 |
ENST00000367157.3
|
NUAK2
|
NUAK family, SNF1-like kinase, 2 |
| chr6_-_31619742 | 0.39 |
ENST00000433828.1
ENST00000456286.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr5_+_150040403 | 0.39 |
ENST00000517768.1
ENST00000297130.4 |
MYOZ3
|
myozenin 3 |
| chr7_-_20826504 | 0.38 |
ENST00000418710.2
ENST00000361443.4 |
SP8
|
Sp8 transcription factor |
| chr7_-_87849340 | 0.38 |
ENST00000419179.1
ENST00000265729.2 |
SRI
|
sorcin |
| chr3_-_120170052 | 0.38 |
ENST00000295633.3
|
FSTL1
|
follistatin-like 1 |
| chr1_+_144989309 | 0.37 |
ENST00000596396.1
|
AL590452.1
|
Uncharacterized protein |
| chr12_-_52685312 | 0.36 |
ENST00000327741.5
|
KRT81
|
keratin 81 |
| chr22_-_31688381 | 0.36 |
ENST00000487265.2
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr17_-_56609302 | 0.36 |
ENST00000581607.1
ENST00000317256.6 ENST00000426861.1 ENST00000580809.1 ENST00000577729.1 ENST00000583291.1 |
SEPT4
|
septin 4 |
| chr11_+_18477369 | 0.35 |
ENST00000396213.3
ENST00000280706.2 |
LDHAL6A
|
lactate dehydrogenase A-like 6A |
| chr17_-_34122596 | 0.35 |
ENST00000250144.8
|
MMP28
|
matrix metallopeptidase 28 |
| chr11_+_120081475 | 0.35 |
ENST00000328965.4
|
OAF
|
OAF homolog (Drosophila) |
| chr19_-_44174330 | 0.35 |
ENST00000340093.3
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr14_-_62217779 | 0.34 |
ENST00000554254.1
|
HIF1A-AS2
|
HIF1A antisense RNA 2 |
| chr8_-_61193947 | 0.33 |
ENST00000317995.4
|
CA8
|
carbonic anhydrase VIII |
| chr19_-_45748628 | 0.33 |
ENST00000590022.1
|
AC006126.4
|
AC006126.4 |
| chr19_-_44174305 | 0.32 |
ENST00000601723.1
ENST00000339082.3 |
PLAUR
|
plasminogen activator, urokinase receptor |
| chr17_-_47925379 | 0.32 |
ENST00000352793.2
ENST00000334568.4 ENST00000398154.1 ENST00000436235.1 ENST00000326219.5 |
TAC4
|
tachykinin 4 (hemokinin) |
| chr1_-_52456352 | 0.32 |
ENST00000371655.3
|
RAB3B
|
RAB3B, member RAS oncogene family |
| chr3_+_193809263 | 0.31 |
ENST00000457815.1
|
RP11-135A1.3
|
RP11-135A1.3 |
| chr7_-_27135591 | 0.31 |
ENST00000343060.4
ENST00000355633.5 |
HOXA1
|
homeobox A1 |
| chr11_-_47616210 | 0.31 |
ENST00000302514.3
|
C1QTNF4
|
C1q and tumor necrosis factor related protein 4 |
| chr6_-_31620095 | 0.31 |
ENST00000424176.1
ENST00000456622.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr12_-_52761262 | 0.31 |
ENST00000257901.3
|
KRT85
|
keratin 85 |
| chr3_-_139195350 | 0.31 |
ENST00000232217.2
|
RBP2
|
retinol binding protein 2, cellular |
| chr12_-_26278030 | 0.30 |
ENST00000242728.4
|
BHLHE41
|
basic helix-loop-helix family, member e41 |
| chr17_-_76356148 | 0.30 |
ENST00000587578.1
ENST00000330871.2 |
SOCS3
|
suppressor of cytokine signaling 3 |
| chr3_+_171844762 | 0.29 |
ENST00000443501.1
|
FNDC3B
|
fibronectin type III domain containing 3B |
| chr16_-_67965756 | 0.29 |
ENST00000571044.1
ENST00000571605.1 |
CTRL
|
chymotrypsin-like |
| chr8_+_1993173 | 0.28 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr4_-_177116772 | 0.28 |
ENST00000280191.2
|
SPATA4
|
spermatogenesis associated 4 |
| chr6_+_43603552 | 0.28 |
ENST00000372171.4
|
MAD2L1BP
|
MAD2L1 binding protein |
| chr1_+_199996733 | 0.28 |
ENST00000236914.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr1_+_89829610 | 0.28 |
ENST00000370456.4
ENST00000535065.1 |
GBP6
|
guanylate binding protein family, member 6 |
| chrX_-_19689106 | 0.28 |
ENST00000379716.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr5_-_119669160 | 0.28 |
ENST00000514240.1
|
CTC-552D5.1
|
CTC-552D5.1 |
| chr3_+_107243204 | 0.27 |
ENST00000456817.1
ENST00000458458.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr3_-_107777208 | 0.27 |
ENST00000398258.3
|
CD47
|
CD47 molecule |
| chr12_+_22778291 | 0.27 |
ENST00000545979.1
|
ETNK1
|
ethanolamine kinase 1 |
| chr4_+_160188889 | 0.26 |
ENST00000264431.4
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr6_-_31619697 | 0.26 |
ENST00000434444.1
|
BAG6
|
BCL2-associated athanogene 6 |
| chr14_-_65569244 | 0.26 |
ENST00000557277.1
ENST00000556892.1 |
MAX
|
MYC associated factor X |
| chr1_+_244227632 | 0.26 |
ENST00000598000.1
|
AL590483.1
|
Uncharacterized protein; cDNA FLJ42623 fis, clone BRACE3015894 |
| chr1_-_46598371 | 0.25 |
ENST00000372006.1
ENST00000425892.1 ENST00000420542.1 ENST00000354242.4 ENST00000340332.6 |
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr13_-_30160925 | 0.25 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr6_-_26124138 | 0.24 |
ENST00000314332.5
ENST00000396984.1 |
HIST1H2BC
|
histone cluster 1, H2bc |
| chr12_-_49351303 | 0.24 |
ENST00000256682.4
|
ARF3
|
ADP-ribosylation factor 3 |
| chr10_+_90660832 | 0.24 |
ENST00000371924.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr6_-_31620149 | 0.24 |
ENST00000435080.1
ENST00000375976.4 ENST00000441054.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr11_+_122709200 | 0.24 |
ENST00000227348.4
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr1_-_85930246 | 0.24 |
ENST00000426972.3
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr17_-_18585131 | 0.24 |
ENST00000443457.1
ENST00000583002.1 |
ZNF286B
|
zinc finger protein 286B |
| chr17_-_76713100 | 0.23 |
ENST00000585509.1
|
CYTH1
|
cytohesin 1 |
| chr17_+_26662679 | 0.23 |
ENST00000578158.1
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr12_-_89746264 | 0.23 |
ENST00000548755.1
|
DUSP6
|
dual specificity phosphatase 6 |
| chr12_+_52695617 | 0.23 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chr1_+_202995611 | 0.23 |
ENST00000367240.2
|
PPFIA4
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 |
| chr19_-_44258770 | 0.23 |
ENST00000601925.1
ENST00000602222.1 ENST00000599804.1 |
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr1_+_145507587 | 0.23 |
ENST00000330165.8
ENST00000369307.3 |
RBM8A
|
RNA binding motif protein 8A |
| chr3_+_177534653 | 0.23 |
ENST00000436078.1
|
RP11-91K9.1
|
RP11-91K9.1 |
| chr11_-_115127611 | 0.23 |
ENST00000545094.1
|
CADM1
|
cell adhesion molecule 1 |
| chr12_-_49351228 | 0.23 |
ENST00000541959.1
ENST00000447318.2 |
ARF3
|
ADP-ribosylation factor 3 |
| chr19_+_48949087 | 0.23 |
ENST00000598711.1
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr7_-_130597935 | 0.22 |
ENST00000447307.1
ENST00000418546.1 |
MIR29B1
|
microRNA 29a |
| chr15_-_80263506 | 0.22 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr2_-_40679186 | 0.22 |
ENST00000406785.2
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr7_+_76101379 | 0.22 |
ENST00000429179.1
|
DTX2
|
deltex homolog 2 (Drosophila) |
| chr16_+_69221028 | 0.22 |
ENST00000336278.4
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2) |
| chr9_-_117267717 | 0.22 |
ENST00000374057.3
|
DFNB31
|
deafness, autosomal recessive 31 |
| chr12_-_89746173 | 0.21 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr17_-_47308128 | 0.21 |
ENST00000413580.1
ENST00000511066.1 |
PHOSPHO1
|
phosphatase, orphan 1 |
| chr19_-_50529193 | 0.21 |
ENST00000596445.1
ENST00000599538.1 |
VRK3
|
vaccinia related kinase 3 |
| chr14_+_91709279 | 0.21 |
ENST00000554096.1
|
CTD-2547L24.3
|
HCG1816139; Uncharacterized protein |
| chr8_-_6914251 | 0.21 |
ENST00000330590.2
|
DEFA5
|
defensin, alpha 5, Paneth cell-specific |
| chr17_-_47308100 | 0.21 |
ENST00000503902.1
ENST00000512250.1 |
PHOSPHO1
|
phosphatase, orphan 1 |
| chrX_-_132549506 | 0.20 |
ENST00000370828.3
|
GPC4
|
glypican 4 |
| chr14_-_51411194 | 0.20 |
ENST00000544180.2
|
PYGL
|
phosphorylase, glycogen, liver |
| chr1_-_46598284 | 0.20 |
ENST00000423209.1
ENST00000262741.5 |
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr2_+_201994042 | 0.20 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chrX_+_102862834 | 0.19 |
ENST00000372627.5
ENST00000243286.3 |
TCEAL3
|
transcription elongation factor A (SII)-like 3 |
| chr11_+_82783097 | 0.19 |
ENST00000501011.2
ENST00000527627.1 ENST00000526795.1 ENST00000533528.1 ENST00000533708.1 ENST00000534499.1 |
RAB30-AS1
|
RAB30 antisense RNA 1 (head to head) |
| chr2_+_201994208 | 0.19 |
ENST00000440180.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr6_-_31619892 | 0.19 |
ENST00000454165.1
ENST00000428326.1 ENST00000452994.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr11_-_74204692 | 0.18 |
ENST00000528085.1
|
LIPT2
|
lipoyl(octanoyl) transferase 2 (putative) |
| chr19_+_55987998 | 0.18 |
ENST00000591164.1
|
ZNF628
|
zinc finger protein 628 |
| chr16_+_75690093 | 0.18 |
ENST00000564671.2
|
TERF2IP
|
telomeric repeat binding factor 2, interacting protein |
| chr18_-_6928495 | 0.18 |
ENST00000580197.1
|
LINC00668
|
long intergenic non-protein coding RNA 668 |
| chr17_-_42452063 | 0.18 |
ENST00000588098.1
|
ITGA2B
|
integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) |
| chr17_-_37557846 | 0.18 |
ENST00000394294.3
ENST00000583610.1 ENST00000264658.6 |
FBXL20
|
F-box and leucine-rich repeat protein 20 |
| chr15_+_60296421 | 0.18 |
ENST00000396057.4
|
FOXB1
|
forkhead box B1 |
| chr2_+_210636697 | 0.18 |
ENST00000439458.1
ENST00000272845.6 |
UNC80
|
unc-80 homolog (C. elegans) |
| chr19_+_1000418 | 0.17 |
ENST00000234389.3
|
GRIN3B
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3B |
| chr17_+_57970469 | 0.17 |
ENST00000443572.2
ENST00000406116.3 ENST00000225577.4 ENST00000393021.3 |
RPS6KB1
|
ribosomal protein S6 kinase, 70kDa, polypeptide 1 |
| chr3_-_126373929 | 0.17 |
ENST00000523403.1
ENST00000524230.2 |
TXNRD3
|
thioredoxin reductase 3 |
| chr1_+_213123976 | 0.17 |
ENST00000366965.2
ENST00000366967.2 |
VASH2
|
vasohibin 2 |
| chrX_+_47050798 | 0.17 |
ENST00000412206.1
ENST00000427561.1 |
UBA1
|
ubiquitin-like modifier activating enzyme 1 |
| chr11_+_66824346 | 0.17 |
ENST00000532559.1
|
RHOD
|
ras homolog family member D |
| chr12_-_70004942 | 0.16 |
ENST00000361484.3
|
LRRC10
|
leucine rich repeat containing 10 |
| chr11_+_107799118 | 0.16 |
ENST00000320578.2
|
RAB39A
|
RAB39A, member RAS oncogene family |
| chr19_+_36236491 | 0.16 |
ENST00000591949.1
|
PSENEN
|
presenilin enhancer gamma secretase subunit |
| chr22_+_42196666 | 0.16 |
ENST00000402061.3
ENST00000255784.5 |
CCDC134
|
coiled-coil domain containing 134 |
| chr15_+_59908633 | 0.16 |
ENST00000559626.1
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr4_-_185303418 | 0.16 |
ENST00000610223.1
ENST00000608785.1 |
RP11-290F5.1
|
RP11-290F5.1 |
| chr1_+_200993071 | 0.16 |
ENST00000446333.1
ENST00000458003.1 |
RP11-168O16.1
|
RP11-168O16.1 |
| chr8_+_38965048 | 0.16 |
ENST00000399831.3
ENST00000437682.2 ENST00000519315.1 ENST00000379907.4 ENST00000522506.1 |
ADAM32
|
ADAM metallopeptidase domain 32 |
| chr18_-_32957260 | 0.16 |
ENST00000587422.1
ENST00000306346.1 ENST00000589332.1 ENST00000586687.1 ENST00000585522.1 |
ZNF396
|
zinc finger protein 396 |
| chr22_+_18560743 | 0.16 |
ENST00000399744.3
|
PEX26
|
peroxisomal biogenesis factor 26 |
| chr3_+_119217422 | 0.16 |
ENST00000466984.1
|
TIMMDC1
|
translocase of inner mitochondrial membrane domain containing 1 |
| chr17_+_25958174 | 0.16 |
ENST00000313648.6
ENST00000577392.1 ENST00000584661.1 ENST00000413914.2 |
LGALS9
|
lectin, galactoside-binding, soluble, 9 |
| chr19_+_37407212 | 0.16 |
ENST00000427117.1
ENST00000587130.1 ENST00000333987.7 ENST00000415168.1 ENST00000444991.1 |
ZNF568
|
zinc finger protein 568 |
| chr1_+_40840320 | 0.16 |
ENST00000372708.1
|
SMAP2
|
small ArfGAP2 |
| chr1_+_20915409 | 0.16 |
ENST00000375071.3
|
CDA
|
cytidine deaminase |
| chr6_+_45389893 | 0.16 |
ENST00000371432.3
|
RUNX2
|
runt-related transcription factor 2 |
| chr19_-_10426663 | 0.16 |
ENST00000541276.1
ENST00000393708.3 ENST00000494368.1 |
FDX1L
|
ferredoxin 1-like |
| chr1_+_41445413 | 0.16 |
ENST00000541520.1
|
CTPS1
|
CTP synthase 1 |
| chr11_-_60010556 | 0.16 |
ENST00000427611.2
|
MS4A4E
|
membrane-spanning 4-domains, subfamily A, member 4E |
| chr6_+_131894284 | 0.15 |
ENST00000368087.3
ENST00000356962.2 |
ARG1
|
arginase 1 |
| chr15_+_41913690 | 0.15 |
ENST00000563576.1
|
MGA
|
MGA, MAX dimerization protein |
| chr19_+_48949030 | 0.15 |
ENST00000253237.5
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr16_+_57844549 | 0.15 |
ENST00000564282.1
|
CTD-2600O9.1
|
uncharacterized protein LOC388282 |
| chr3_+_57741957 | 0.15 |
ENST00000295951.3
|
SLMAP
|
sarcolemma associated protein |
| chr19_-_44258733 | 0.15 |
ENST00000597586.1
ENST00000596714.1 |
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr6_-_154751629 | 0.15 |
ENST00000424998.1
|
CNKSR3
|
CNKSR family member 3 |
| chr16_-_79634595 | 0.15 |
ENST00000326043.4
ENST00000393350.1 |
MAF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr22_-_38699003 | 0.15 |
ENST00000451964.1
|
CSNK1E
|
casein kinase 1, epsilon |
| chr2_-_61697862 | 0.15 |
ENST00000398571.2
|
USP34
|
ubiquitin specific peptidase 34 |
| chr13_+_46276441 | 0.15 |
ENST00000310521.1
ENST00000533564.1 |
SPERT
|
spermatid associated |
| chr8_+_145218673 | 0.15 |
ENST00000326134.5
|
MROH1
|
maestro heat-like repeat family member 1 |
| chr1_+_155294342 | 0.15 |
ENST00000292254.4
|
RUSC1
|
RUN and SH3 domain containing 1 |
| chr4_-_89619386 | 0.15 |
ENST00000323061.5
|
NAP1L5
|
nucleosome assembly protein 1-like 5 |
| chr19_-_13213662 | 0.15 |
ENST00000264824.4
|
LYL1
|
lymphoblastic leukemia derived sequence 1 |
| chr2_-_177684007 | 0.15 |
ENST00000451851.1
|
AC092162.1
|
AC092162.1 |
| chr17_-_79604075 | 0.15 |
ENST00000374747.5
ENST00000539314.1 ENST00000331134.6 |
NPLOC4
|
nuclear protein localization 4 homolog (S. cerevisiae) |
| chr17_+_7358889 | 0.14 |
ENST00000575379.1
|
CHRNB1
|
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr20_+_30193083 | 0.14 |
ENST00000376112.3
ENST00000376105.3 |
ID1
|
inhibitor of DNA binding 1, dominant negative helix-loop-helix protein |
| chr2_-_228497888 | 0.14 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chr1_-_19229218 | 0.14 |
ENST00000432718.1
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr2_-_113594279 | 0.14 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr8_+_145490549 | 0.14 |
ENST00000340695.2
|
SCXA
|
scleraxis homolog A (mouse) |
| chr1_+_27648648 | 0.14 |
ENST00000374076.4
|
TMEM222
|
transmembrane protein 222 |
| chr2_-_200322723 | 0.14 |
ENST00000417098.1
|
SATB2
|
SATB homeobox 2 |
| chr22_-_42915788 | 0.14 |
ENST00000323013.6
|
RRP7A
|
ribosomal RNA processing 7 homolog A (S. cerevisiae) |
| chr11_-_111649074 | 0.14 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr6_+_26383318 | 0.14 |
ENST00000469230.1
ENST00000490025.1 ENST00000356709.4 ENST00000352867.2 ENST00000493275.1 ENST00000472507.1 ENST00000482536.1 ENST00000432533.2 ENST00000482842.1 |
BTN2A2
|
butyrophilin, subfamily 2, member A2 |
| chr2_-_74776586 | 0.14 |
ENST00000420535.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr7_+_150076406 | 0.14 |
ENST00000329630.5
|
ZNF775
|
zinc finger protein 775 |
| chr15_-_38852251 | 0.14 |
ENST00000558432.1
|
RASGRP1
|
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
| chr15_-_52030293 | 0.14 |
ENST00000560491.1
ENST00000267838.3 |
LYSMD2
|
LysM, putative peptidoglycan-binding, domain containing 2 |
| chr20_-_1165319 | 0.14 |
ENST00000429036.1
|
TMEM74B
|
transmembrane protein 74B |
| chrX_-_69509738 | 0.14 |
ENST00000374454.1
ENST00000239666.4 |
PDZD11
|
PDZ domain containing 11 |
| chr3_+_121554046 | 0.14 |
ENST00000273668.2
ENST00000451944.2 |
EAF2
|
ELL associated factor 2 |
| chr14_+_23727694 | 0.14 |
ENST00000399905.1
ENST00000470456.1 |
C14orf164
|
chromosome 14 open reading frame 164 |
| chr14_-_51411146 | 0.14 |
ENST00000532462.1
|
PYGL
|
phosphorylase, glycogen, liver |
| chr21_+_30503282 | 0.13 |
ENST00000399925.1
|
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr17_-_40428359 | 0.13 |
ENST00000293328.3
|
STAT5B
|
signal transducer and activator of transcription 5B |
| chr14_-_81687575 | 0.13 |
ENST00000434192.2
|
GTF2A1
|
general transcription factor IIA, 1, 19/37kDa |
| chr2_+_228678550 | 0.13 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr9_-_86322516 | 0.13 |
ENST00000529923.1
|
UBQLN1
|
ubiquilin 1 |
| chr16_+_77233294 | 0.13 |
ENST00000378644.4
|
SYCE1L
|
synaptonemal complex central element protein 1-like |
| chr9_+_22646189 | 0.13 |
ENST00000436786.1
|
RP11-399D6.2
|
RP11-399D6.2 |
| chr16_+_72459838 | 0.13 |
ENST00000564508.1
|
AC004158.3
|
AC004158.3 |
| chr1_+_26737253 | 0.13 |
ENST00000326279.6
|
LIN28A
|
lin-28 homolog A (C. elegans) |
| chr17_+_46184911 | 0.13 |
ENST00000580219.1
ENST00000452859.2 ENST00000393405.2 ENST00000439357.2 ENST00000359238.2 |
SNX11
|
sorting nexin 11 |
| chr15_-_83316087 | 0.13 |
ENST00000568757.1
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr3_-_196065248 | 0.13 |
ENST00000446879.1
ENST00000273695.3 |
TM4SF19
|
transmembrane 4 L six family member 19 |
| chr15_-_33180439 | 0.13 |
ENST00000559610.1
|
FMN1
|
formin 1 |
| chr4_-_90756769 | 0.13 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX21_TBR1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.3 | 0.8 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.2 | 0.8 | GO:0001757 | somite specification(GO:0001757) |
| 0.2 | 0.9 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.2 | 0.5 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 0.7 | GO:0001315 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 1.4 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.1 | 0.5 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.1 | 0.6 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.1 | 0.5 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.1 | 0.3 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.1 | 0.2 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) |
| 0.1 | 0.2 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 | 0.7 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.1 | 0.8 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.1 | 0.4 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 0.4 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.1 | 0.4 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 | 0.3 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 0.4 | GO:0045354 | negative regulation of interferon-alpha production(GO:0032687) interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.1 | 0.4 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.3 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.1 | 0.3 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 0.3 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.2 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.2 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.0 | 0.3 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 0.0 | 0.4 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) negative regulation of heart rate(GO:0010459) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.3 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.4 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.2 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0045362 | interleukin-1 biosynthetic process(GO:0042222) regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.0 | 0.1 | GO:1901340 | negative regulation of store-operated calcium channel activity(GO:1901340) |
| 0.0 | 0.2 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.2 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.2 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.2 | GO:0043091 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.0 | 0.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.4 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.2 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.2 | GO:0051621 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.5 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.1 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.4 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.2 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.2 | GO:0002860 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.4 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.2 | GO:0046717 | glutamate secretion(GO:0014047) acid secretion(GO:0046717) |
| 0.0 | 0.1 | GO:2000329 | peptidyl-lysine oxidation(GO:0018057) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.3 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:2000987 | hormone-mediated apoptotic signaling pathway(GO:0008628) cellular response to cocaine(GO:0071314) positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.1 | GO:0032826 | natural killer cell differentiation involved in immune response(GO:0002325) regulation of natural killer cell differentiation involved in immune response(GO:0032826) regulation of cytolysis in other organism(GO:0051710) |
| 0.0 | 0.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.0 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 0.2 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.3 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.1 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.2 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 0.0 | 0.1 | GO:1903026 | negative regulation of endothelial cell differentiation(GO:0045602) negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.6 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 | 0.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.0 | 0.1 | GO:1905066 | regulation of Wnt signaling pathway involved in heart development(GO:0003307) arterial endothelial cell fate commitment(GO:0060844) positive regulation of ephrin receptor signaling pathway(GO:1901189) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.1 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.1 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.4 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.0 | GO:0032661 | central nervous system myelin formation(GO:0032289) regulation of interleukin-18 production(GO:0032661) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.1 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.2 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.0 | 0.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.0 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.1 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.5 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.0 | 0.3 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.1 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.0 | 0.1 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.0 | 0.1 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.1 | GO:0043622 | cortical microtubule organization(GO:0043622) establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.0 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.2 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.1 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.1 | GO:0035864 | response to potassium ion(GO:0035864) |
| 0.0 | 0.2 | GO:1904871 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.3 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.0 | 0.3 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.1 | 1.4 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 0.4 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.1 | 0.4 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.7 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.4 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.1 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.0 | 0.1 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.0 | 0.4 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.4 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.0 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.3 | 0.9 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.2 | 0.5 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.4 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.7 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.1 | 0.7 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.3 | GO:0008184 | purine nucleobase binding(GO:0002060) glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.5 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 1.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.2 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.1 | 0.6 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.2 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.2 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.2 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.1 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.3 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.3 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.2 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.4 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.2 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.4 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.8 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.0 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 0.0 | 0.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.2 | GO:1990825 | G-quadruplex RNA binding(GO:0002151) sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.2 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.5 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.7 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.4 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.0 | GO:0004461 | lactose synthase activity(GO:0004461) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.3 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.5 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.6 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.8 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.3 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.9 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.4 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.8 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.5 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.7 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |