Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for TBX20
Z-value: 0.66
Transcription factors associated with TBX20
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX20
|
ENSG00000164532.10 | T-box transcription factor 20 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX20 | hg19_v2_chr7_-_35293740_35293758 | 0.33 | 6.7e-01 | Click! |
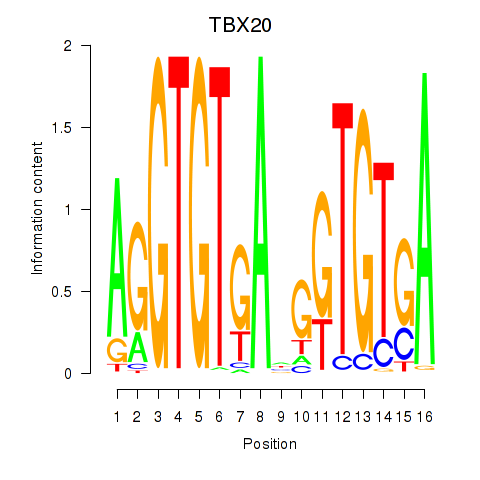
Activity profile of TBX20 motif
Sorted Z-values of TBX20 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_57662527 | 1.72 |
ENST00000563374.1
ENST00000568234.1 ENST00000565770.1 ENST00000564338.1 ENST00000566164.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr16_+_57662419 | 1.11 |
ENST00000388812.4
ENST00000538815.1 ENST00000456916.1 ENST00000567154.1 ENST00000388813.5 ENST00000562558.1 ENST00000566271.2 |
GPR56
|
G protein-coupled receptor 56 |
| chr19_+_56915668 | 0.86 |
ENST00000333201.9
ENST00000391778.3 |
ZNF583
|
zinc finger protein 583 |
| chr8_+_142264664 | 0.56 |
ENST00000518520.1
|
RP11-10J21.3
|
Uncharacterized protein |
| chr2_+_64068116 | 0.46 |
ENST00000480679.1
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr15_+_76016293 | 0.33 |
ENST00000332145.2
|
ODF3L1
|
outer dense fiber of sperm tails 3-like 1 |
| chr17_-_7080227 | 0.31 |
ENST00000574330.1
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr11_-_14358620 | 0.28 |
ENST00000531421.1
|
RRAS2
|
related RAS viral (r-ras) oncogene homolog 2 |
| chr2_-_219134343 | 0.27 |
ENST00000447885.1
ENST00000420660.1 |
AAMP
|
angio-associated, migratory cell protein |
| chr2_+_64068074 | 0.27 |
ENST00000394417.2
ENST00000484142.1 ENST00000482668.1 ENST00000467648.2 |
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr3_-_139396801 | 0.27 |
ENST00000413939.2
ENST00000339837.5 ENST00000512391.1 |
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr1_-_154909329 | 0.20 |
ENST00000368467.3
|
PMVK
|
phosphomevalonate kinase |
| chr19_+_35810164 | 0.20 |
ENST00000598537.1
|
CD22
|
CD22 molecule |
| chr3_-_139396853 | 0.19 |
ENST00000406164.1
ENST00000406824.1 |
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr2_-_219134822 | 0.18 |
ENST00000444053.1
ENST00000248450.4 |
AAMP
|
angio-associated, migratory cell protein |
| chr13_+_113760098 | 0.17 |
ENST00000346342.3
ENST00000541084.1 ENST00000375581.3 |
F7
|
coagulation factor VII (serum prothrombin conversion accelerator) |
| chr3_-_15140629 | 0.16 |
ENST00000507357.1
ENST00000449050.1 ENST00000253699.3 ENST00000435849.3 ENST00000476527.2 |
ZFYVE20
|
zinc finger, FYVE domain containing 20 |
| chr2_+_207630081 | 0.15 |
ENST00000236980.6
ENST00000418289.1 ENST00000402774.3 ENST00000403094.3 |
FASTKD2
|
FAST kinase domains 2 |
| chr2_-_207629997 | 0.14 |
ENST00000454776.2
|
MDH1B
|
malate dehydrogenase 1B, NAD (soluble) |
| chr20_-_48782639 | 0.14 |
ENST00000435301.2
|
RP11-112L6.3
|
RP11-112L6.3 |
| chr2_+_173940442 | 0.14 |
ENST00000409176.2
ENST00000338983.3 ENST00000431503.2 |
MLTK
|
Mitogen-activated protein kinase kinase kinase MLT |
| chr15_-_74421477 | 0.13 |
ENST00000514871.1
|
RP11-247C2.2
|
HCG2004779; Uncharacterized protein |
| chr12_-_56123444 | 0.10 |
ENST00000546457.1
ENST00000549117.1 |
CD63
|
CD63 molecule |
| chr5_-_89825328 | 0.09 |
ENST00000500869.2
ENST00000315948.6 ENST00000509384.1 |
LYSMD3
|
LysM, putative peptidoglycan-binding, domain containing 3 |
| chr10_+_114135952 | 0.08 |
ENST00000356116.1
ENST00000433418.1 ENST00000354273.4 |
ACSL5
|
acyl-CoA synthetase long-chain family member 5 |
| chr2_-_207630033 | 0.08 |
ENST00000449792.1
|
MDH1B
|
malate dehydrogenase 1B, NAD (soluble) |
| chr1_+_43824669 | 0.07 |
ENST00000372462.1
|
CDC20
|
cell division cycle 20 |
| chr5_+_133861339 | 0.07 |
ENST00000282605.4
ENST00000361895.2 ENST00000402835.1 |
PHF15
|
jade family PHD finger 2 |
| chr17_-_3599492 | 0.07 |
ENST00000435558.1
ENST00000345901.3 |
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr19_+_44331555 | 0.07 |
ENST00000590950.1
|
ZNF283
|
zinc finger protein 283 |
| chr1_-_17380630 | 0.06 |
ENST00000375499.3
|
SDHB
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr2_+_61108650 | 0.05 |
ENST00000295025.8
|
REL
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr19_+_46171464 | 0.05 |
ENST00000590918.1
ENST00000263281.3 ENST00000304207.8 |
GIPR
|
gastric inhibitory polypeptide receptor |
| chr5_+_135394840 | 0.05 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr3_+_142315225 | 0.05 |
ENST00000457734.2
ENST00000483373.1 ENST00000475296.1 ENST00000495744.1 ENST00000476044.1 ENST00000461644.1 |
PLS1
|
plastin 1 |
| chr19_-_4540486 | 0.05 |
ENST00000306390.6
|
LRG1
|
leucine-rich alpha-2-glycoprotein 1 |
| chrX_+_67913471 | 0.05 |
ENST00000374597.3
|
STARD8
|
StAR-related lipid transfer (START) domain containing 8 |
| chr2_+_176972000 | 0.05 |
ENST00000249504.5
|
HOXD11
|
homeobox D11 |
| chr20_+_44509857 | 0.04 |
ENST00000372523.1
ENST00000372520.1 |
ZSWIM1
|
zinc finger, SWIM-type containing 1 |
| chr22_+_38219389 | 0.03 |
ENST00000249041.2
|
GALR3
|
galanin receptor 3 |
| chr10_+_114135004 | 0.03 |
ENST00000393081.1
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5 |
| chr6_-_2842087 | 0.03 |
ENST00000537185.1
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr7_+_30185406 | 0.03 |
ENST00000324489.5
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr1_+_162760513 | 0.02 |
ENST00000367915.1
ENST00000367917.3 ENST00000254521.3 ENST00000367913.1 |
HSD17B7
|
hydroxysteroid (17-beta) dehydrogenase 7 |
| chr2_+_85360499 | 0.02 |
ENST00000282111.3
|
TCF7L1
|
transcription factor 7-like 1 (T-cell specific, HMG-box) |
| chr6_-_2842219 | 0.02 |
ENST00000380739.5
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr17_-_3599696 | 0.01 |
ENST00000225328.5
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr7_+_30185496 | 0.01 |
ENST00000455738.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr3_-_131221771 | 0.01 |
ENST00000510154.1
ENST00000507669.1 |
MRPL3
|
mitochondrial ribosomal protein L3 |
| chr14_+_77228532 | 0.01 |
ENST00000167106.4
ENST00000554237.1 |
VASH1
|
vasohibin 1 |
| chr12_-_16762971 | 0.00 |
ENST00000540590.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX20
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.1 | 0.5 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.2 | GO:1905225 | response to thyrotropin-releasing hormone(GO:1905225) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.1 | 0.5 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.3 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |