Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
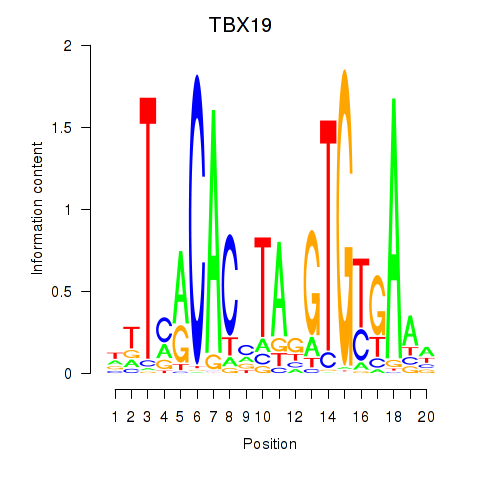
Results for TBX19
Z-value: 1.43
Transcription factors associated with TBX19
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX19
|
ENSG00000143178.8 | T-box transcription factor 19 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX19 | hg19_v2_chr1_+_168250194_168250278 | -0.36 | 6.4e-01 | Click! |
Activity profile of TBX19 motif
Sorted Z-values of TBX19 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_150866779 | 3.29 |
ENST00000370353.3
|
PRRG3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr10_+_43916052 | 2.26 |
ENST00000442526.2
|
RP11-517P14.2
|
RP11-517P14.2 |
| chr9_+_78505581 | 1.20 |
ENST00000376767.3
ENST00000376752.4 |
PCSK5
|
proprotein convertase subtilisin/kexin type 5 |
| chr1_+_100598691 | 1.17 |
ENST00000370143.1
ENST00000370141.2 |
TRMT13
|
tRNA methyltransferase 13 homolog (S. cerevisiae) |
| chr3_+_98482175 | 0.93 |
ENST00000485391.1
ENST00000492254.1 |
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr19_-_49568311 | 0.84 |
ENST00000595857.1
ENST00000451356.2 |
NTF4
|
neurotrophin 4 |
| chr1_-_63988846 | 0.76 |
ENST00000283568.8
ENST00000371092.3 ENST00000271002.10 |
ITGB3BP
|
integrin beta 3 binding protein (beta3-endonexin) |
| chr2_-_201753859 | 0.76 |
ENST00000409361.1
ENST00000392283.4 |
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr5_-_159846066 | 0.75 |
ENST00000519349.1
ENST00000520664.1 |
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae) |
| chr6_+_26204825 | 0.75 |
ENST00000360441.4
|
HIST1H4E
|
histone cluster 1, H4e |
| chr21_+_39628655 | 0.73 |
ENST00000398925.1
ENST00000398928.1 ENST00000328656.4 ENST00000443341.1 |
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr14_+_105212297 | 0.71 |
ENST00000556623.1
ENST00000555674.1 |
ADSSL1
|
adenylosuccinate synthase like 1 |
| chr14_+_105147464 | 0.68 |
ENST00000540171.2
|
RP11-982M15.6
|
RP11-982M15.6 |
| chr10_+_90750378 | 0.65 |
ENST00000355740.2
ENST00000352159.4 |
FAS
|
Fas cell surface death receptor |
| chr6_-_38607673 | 0.63 |
ENST00000481247.1
|
BTBD9
|
BTB (POZ) domain containing 9 |
| chr14_+_50159813 | 0.60 |
ENST00000359332.2
ENST00000553274.1 ENST00000557128.1 |
KLHDC1
|
kelch domain containing 1 |
| chrX_-_119709637 | 0.59 |
ENST00000404115.3
|
CUL4B
|
cullin 4B |
| chr2_-_201753717 | 0.58 |
ENST00000409264.2
|
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr7_+_47834880 | 0.58 |
ENST00000258776.4
|
C7orf69
|
chromosome 7 open reading frame 69 |
| chr9_+_130890612 | 0.55 |
ENST00000443493.1
|
AL590708.2
|
AL590708.2 |
| chrX_-_15402498 | 0.54 |
ENST00000297904.3
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D) |
| chr5_-_78281623 | 0.54 |
ENST00000521117.1
|
ARSB
|
arylsulfatase B |
| chr20_-_3762087 | 0.50 |
ENST00000379756.3
|
SPEF1
|
sperm flagellar 1 |
| chr6_+_153552455 | 0.49 |
ENST00000392385.2
|
AL590867.1
|
Uncharacterized protein; cDNA FLJ59044, highly similar to LINE-1 reverse transcriptase homolog |
| chr8_-_117886563 | 0.49 |
ENST00000519837.1
ENST00000522699.1 |
RAD21
|
RAD21 homolog (S. pombe) |
| chr8_+_94767072 | 0.48 |
ENST00000452276.1
ENST00000453321.3 ENST00000498673.1 ENST00000518319.1 |
TMEM67
|
transmembrane protein 67 |
| chr1_-_167522982 | 0.46 |
ENST00000370509.4
|
CREG1
|
cellular repressor of E1A-stimulated genes 1 |
| chr17_+_6659354 | 0.45 |
ENST00000574907.1
|
XAF1
|
XIAP associated factor 1 |
| chr2_-_201753980 | 0.45 |
ENST00000443398.1
ENST00000286175.8 ENST00000409449.1 |
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr6_+_123110465 | 0.44 |
ENST00000539041.1
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr1_-_19615744 | 0.41 |
ENST00000361640.4
|
AKR7A3
|
aldo-keto reductase family 7, member A3 (aflatoxin aldehyde reductase) |
| chr14_+_32546145 | 0.41 |
ENST00000556611.1
ENST00000539826.2 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr6_+_53794780 | 0.40 |
ENST00000505762.1
ENST00000511369.1 ENST00000431554.2 |
MLIP
RP11-411K7.1
|
muscular LMNA-interacting protein RP11-411K7.1 |
| chr15_-_74494779 | 0.40 |
ENST00000571341.1
|
STRA6
|
stimulated by retinoic acid 6 |
| chr8_+_94767109 | 0.40 |
ENST00000409623.3
ENST00000453906.1 ENST00000521517.1 |
TMEM67
|
transmembrane protein 67 |
| chr19_+_21203426 | 0.39 |
ENST00000261560.5
ENST00000599548.1 ENST00000594110.1 |
ZNF430
|
zinc finger protein 430 |
| chr2_+_201754135 | 0.39 |
ENST00000409357.1
ENST00000409129.2 |
NIF3L1
|
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr6_+_52285046 | 0.38 |
ENST00000371068.5
|
EFHC1
|
EF-hand domain (C-terminal) containing 1 |
| chr3_+_98482159 | 0.38 |
ENST00000468553.1
|
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr15_+_63889577 | 0.38 |
ENST00000534939.1
ENST00000539570.3 |
FBXL22
|
F-box and leucine-rich repeat protein 22 |
| chr1_+_236557569 | 0.37 |
ENST00000334232.4
|
EDARADD
|
EDAR-associated death domain |
| chr6_+_26217159 | 0.37 |
ENST00000303910.2
|
HIST1H2AE
|
histone cluster 1, H2ae |
| chr4_+_15704679 | 0.36 |
ENST00000382346.3
|
BST1
|
bone marrow stromal cell antigen 1 |
| chr10_+_95653687 | 0.36 |
ENST00000371408.3
ENST00000427197.1 |
SLC35G1
|
solute carrier family 35, member G1 |
| chr2_+_201754050 | 0.36 |
ENST00000426253.1
ENST00000416651.1 ENST00000454952.1 ENST00000409020.1 ENST00000359683.4 |
NIF3L1
|
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr19_+_852291 | 0.35 |
ENST00000263621.1
|
ELANE
|
elastase, neutrophil expressed |
| chr3_-_123680047 | 0.35 |
ENST00000409697.3
|
CCDC14
|
coiled-coil domain containing 14 |
| chr8_-_19540086 | 0.35 |
ENST00000332246.6
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr19_+_58514229 | 0.35 |
ENST00000546949.1
ENST00000553254.1 ENST00000547364.1 |
CTD-2368P22.1
|
HCG1811579; Uncharacterized protein |
| chr8_-_19540266 | 0.35 |
ENST00000311540.4
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr3_+_108308845 | 0.34 |
ENST00000479138.1
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr1_-_145382362 | 0.33 |
ENST00000419817.1
ENST00000421937.3 ENST00000433081.2 |
RP11-458D21.1
|
RP11-458D21.1 |
| chr4_+_123653807 | 0.33 |
ENST00000314218.3
ENST00000542236.1 |
BBS12
|
Bardet-Biedl syndrome 12 |
| chr1_-_100715372 | 0.33 |
ENST00000370131.3
ENST00000370132.4 |
DBT
|
dihydrolipoamide branched chain transacylase E2 |
| chr14_+_69658194 | 0.33 |
ENST00000409018.3
ENST00000409014.1 ENST00000409675.1 |
EXD2
|
exonuclease 3'-5' domain containing 2 |
| chr15_-_65809581 | 0.32 |
ENST00000341861.5
|
DPP8
|
dipeptidyl-peptidase 8 |
| chr16_-_18937072 | 0.31 |
ENST00000569122.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr4_+_71600063 | 0.30 |
ENST00000513597.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chrX_-_138914394 | 0.30 |
ENST00000327569.3
ENST00000361648.2 ENST00000370543.1 ENST00000359686.2 |
ATP11C
|
ATPase, class VI, type 11C |
| chr20_-_5093673 | 0.30 |
ENST00000379299.2
ENST00000379286.2 ENST00000379279.2 ENST00000379283.2 |
TMEM230
|
transmembrane protein 230 |
| chr11_-_59436453 | 0.29 |
ENST00000300146.9
|
PATL1
|
protein associated with topoisomerase II homolog 1 (yeast) |
| chr5_-_76383133 | 0.29 |
ENST00000255198.2
|
ZBED3
|
zinc finger, BED-type containing 3 |
| chr9_-_116172946 | 0.28 |
ENST00000374171.4
|
POLE3
|
polymerase (DNA directed), epsilon 3, accessory subunit |
| chr5_+_33440802 | 0.28 |
ENST00000502553.1
ENST00000514259.1 ENST00000265112.3 |
TARS
|
threonyl-tRNA synthetase |
| chr3_+_185431080 | 0.27 |
ENST00000296270.1
|
C3orf65
|
chromosome 3 open reading frame 65 |
| chr12_-_8218997 | 0.27 |
ENST00000307637.4
|
C3AR1
|
complement component 3a receptor 1 |
| chr9_-_117150303 | 0.27 |
ENST00000312033.3
|
AKNA
|
AT-hook transcription factor |
| chr19_+_35168547 | 0.27 |
ENST00000502743.1
ENST00000509528.1 ENST00000506901.1 |
ZNF302
|
zinc finger protein 302 |
| chr1_+_89990431 | 0.26 |
ENST00000330947.2
ENST00000358200.4 |
LRRC8B
|
leucine rich repeat containing 8 family, member B |
| chr15_+_69373210 | 0.26 |
ENST00000435479.1
ENST00000559870.1 |
LINC00277
RP11-809H16.5
|
long intergenic non-protein coding RNA 277 RP11-809H16.5 |
| chr4_+_15704573 | 0.26 |
ENST00000265016.4
|
BST1
|
bone marrow stromal cell antigen 1 |
| chr15_-_60771280 | 0.26 |
ENST00000560072.1
ENST00000560406.1 ENST00000560520.1 ENST00000261520.4 ENST00000439632.1 |
NARG2
|
NMDA receptor regulated 2 |
| chr13_+_21141208 | 0.26 |
ENST00000351808.5
|
IFT88
|
intraflagellar transport 88 homolog (Chlamydomonas) |
| chr2_-_162931052 | 0.26 |
ENST00000360534.3
|
DPP4
|
dipeptidyl-peptidase 4 |
| chr20_-_5093713 | 0.26 |
ENST00000342308.5
ENST00000202834.7 |
TMEM230
|
transmembrane protein 230 |
| chr16_+_20685815 | 0.25 |
ENST00000561584.1
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr5_-_177659539 | 0.25 |
ENST00000476170.2
|
PHYKPL
|
5-phosphohydroxy-L-lysine phospho-lyase |
| chr6_+_26158343 | 0.25 |
ENST00000377777.4
ENST00000289316.2 |
HIST1H2BD
|
histone cluster 1, H2bd |
| chr3_-_141944398 | 0.25 |
ENST00000544571.1
ENST00000392993.2 |
GK5
|
glycerol kinase 5 (putative) |
| chr18_-_19180681 | 0.24 |
ENST00000269214.5
|
ESCO1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr5_-_177659625 | 0.24 |
ENST00000323594.4
|
PHYKPL
|
5-phosphohydroxy-L-lysine phospho-lyase |
| chr8_-_117886612 | 0.24 |
ENST00000520992.1
|
RAD21
|
RAD21 homolog (S. pombe) |
| chr1_+_202091980 | 0.24 |
ENST00000367282.5
|
GPR37L1
|
G protein-coupled receptor 37 like 1 |
| chr17_-_7123021 | 0.24 |
ENST00000399510.2
|
DLG4
|
discs, large homolog 4 (Drosophila) |
| chr14_-_58894332 | 0.23 |
ENST00000395159.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr2_-_9563575 | 0.23 |
ENST00000488451.1
ENST00000238091.4 ENST00000355346.4 |
ITGB1BP1
|
integrin beta 1 binding protein 1 |
| chr1_-_243326612 | 0.23 |
ENST00000492145.1
ENST00000490813.1 ENST00000464936.1 |
CEP170
|
centrosomal protein 170kDa |
| chr3_+_88198838 | 0.23 |
ENST00000318887.3
|
C3orf38
|
chromosome 3 open reading frame 38 |
| chr11_+_1092184 | 0.23 |
ENST00000361558.6
|
MUC2
|
mucin 2, oligomeric mucus/gel-forming |
| chr7_+_76751926 | 0.23 |
ENST00000285871.4
ENST00000431197.1 |
CCDC146
|
coiled-coil domain containing 146 |
| chr1_-_179457805 | 0.23 |
ENST00000600581.1
|
AL160286.1
|
Uncharacterized protein |
| chr5_-_70272055 | 0.23 |
ENST00000514857.2
|
NAIP
|
NLR family, apoptosis inhibitory protein |
| chr6_+_52285131 | 0.22 |
ENST00000433625.2
|
EFHC1
|
EF-hand domain (C-terminal) containing 1 |
| chr6_-_28321827 | 0.22 |
ENST00000444081.1
|
ZSCAN31
|
zinc finger and SCAN domain containing 31 |
| chr7_-_76255444 | 0.22 |
ENST00000454397.1
|
POMZP3
|
POM121 and ZP3 fusion |
| chr5_+_96211643 | 0.22 |
ENST00000437043.3
ENST00000510373.1 |
ERAP2
|
endoplasmic reticulum aminopeptidase 2 |
| chr17_-_1463095 | 0.22 |
ENST00000575895.1
ENST00000573056.1 |
PITPNA
|
phosphatidylinositol transfer protein, alpha |
| chr20_+_43835638 | 0.22 |
ENST00000372781.3
ENST00000244069.6 |
SEMG1
|
semenogelin I |
| chr2_-_211341411 | 0.21 |
ENST00000233714.4
ENST00000443314.1 ENST00000441020.3 ENST00000450366.2 ENST00000431941.2 |
LANCL1
|
LanC lantibiotic synthetase component C-like 1 (bacterial) |
| chr10_+_17686193 | 0.21 |
ENST00000377500.1
|
STAM
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
| chr15_-_43910998 | 0.20 |
ENST00000450892.2
|
STRC
|
stereocilin |
| chr7_+_112120908 | 0.20 |
ENST00000439068.2
ENST00000312849.4 ENST00000429049.1 |
LSMEM1
|
leucine-rich single-pass membrane protein 1 |
| chr1_-_100598444 | 0.20 |
ENST00000535161.1
ENST00000287482.5 |
SASS6
|
spindle assembly 6 homolog (C. elegans) |
| chr14_-_69658127 | 0.20 |
ENST00000556182.1
|
RP11-363J20.1
|
RP11-363J20.1 |
| chr2_-_9563216 | 0.19 |
ENST00000467606.1
ENST00000494563.1 ENST00000460001.1 |
ITGB1BP1
|
integrin beta 1 binding protein 1 |
| chr3_+_132316081 | 0.19 |
ENST00000249887.2
|
ACKR4
|
atypical chemokine receptor 4 |
| chr15_-_55581954 | 0.19 |
ENST00000336787.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr19_+_58111241 | 0.19 |
ENST00000597700.1
ENST00000332854.6 ENST00000597864.1 |
ZNF530
|
zinc finger protein 530 |
| chr11_+_30344595 | 0.19 |
ENST00000282032.3
|
ARL14EP
|
ADP-ribosylation factor-like 14 effector protein |
| chr2_-_70409953 | 0.19 |
ENST00000419381.1
|
C2orf42
|
chromosome 2 open reading frame 42 |
| chr5_-_39270725 | 0.19 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr19_+_46003056 | 0.19 |
ENST00000401593.1
ENST00000396736.2 |
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr5_+_98264867 | 0.18 |
ENST00000513175.1
|
CTD-2007H13.3
|
CTD-2007H13.3 |
| chr5_-_110848189 | 0.17 |
ENST00000296632.3
ENST00000512160.1 ENST00000509887.1 |
STARD4
|
StAR-related lipid transfer (START) domain containing 4 |
| chr2_+_120770645 | 0.17 |
ENST00000443902.2
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr2_+_191221240 | 0.17 |
ENST00000409027.1
ENST00000458193.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr10_+_17686221 | 0.17 |
ENST00000540523.1
|
STAM
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
| chr8_+_125954281 | 0.17 |
ENST00000510897.2
ENST00000533286.1 |
LINC00964
|
long intergenic non-protein coding RNA 964 |
| chr6_+_42847649 | 0.16 |
ENST00000424341.2
ENST00000602561.1 |
RPL7L1
|
ribosomal protein L7-like 1 |
| chr2_+_172309634 | 0.16 |
ENST00000339506.3
|
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr8_-_42360015 | 0.16 |
ENST00000522707.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr17_+_29815113 | 0.15 |
ENST00000583755.1
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II) |
| chr19_+_37837185 | 0.15 |
ENST00000541583.2
|
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chr6_-_10694766 | 0.15 |
ENST00000460742.2
ENST00000259983.3 ENST00000379586.1 |
C6orf52
|
chromosome 6 open reading frame 52 |
| chr3_-_179169181 | 0.15 |
ENST00000497513.1
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4 |
| chr4_-_142199943 | 0.14 |
ENST00000514347.1
|
RP11-586L23.1
|
RP11-586L23.1 |
| chr7_-_47578840 | 0.14 |
ENST00000450444.1
|
TNS3
|
tensin 3 |
| chr7_+_48494660 | 0.14 |
ENST00000411975.1
ENST00000544596.1 |
ABCA13
|
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr2_+_228337079 | 0.14 |
ENST00000409315.1
ENST00000373671.3 ENST00000409171.1 |
AGFG1
|
ArfGAP with FG repeats 1 |
| chr2_-_26864228 | 0.14 |
ENST00000288861.4
|
CIB4
|
calcium and integrin binding family member 4 |
| chr9_-_4666421 | 0.14 |
ENST00000381895.5
|
SPATA6L
|
spermatogenesis associated 6-like |
| chr7_-_132766800 | 0.14 |
ENST00000542753.1
ENST00000448878.1 |
CHCHD3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr16_+_31404624 | 0.14 |
ENST00000389202.2
|
ITGAD
|
integrin, alpha D |
| chr12_+_75784850 | 0.14 |
ENST00000550916.1
ENST00000435775.1 ENST00000378689.2 ENST00000378692.3 ENST00000320460.4 ENST00000547164.1 |
GLIPR1L2
|
GLI pathogenesis-related 1 like 2 |
| chr11_+_46740730 | 0.14 |
ENST00000311907.5
ENST00000530231.1 ENST00000442468.1 |
F2
|
coagulation factor II (thrombin) |
| chr10_+_12237924 | 0.13 |
ENST00000429258.2
ENST00000281141.4 |
CDC123
|
cell division cycle 123 |
| chr13_+_100741269 | 0.13 |
ENST00000376286.4
ENST00000376279.3 ENST00000376285.1 |
PCCA
|
propionyl CoA carboxylase, alpha polypeptide |
| chr3_+_32023232 | 0.13 |
ENST00000360311.4
|
ZNF860
|
zinc finger protein 860 |
| chr5_-_159846399 | 0.13 |
ENST00000297151.4
|
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae) |
| chr5_+_140165876 | 0.12 |
ENST00000504120.2
ENST00000394633.3 ENST00000378133.3 |
PCDHA1
|
protocadherin alpha 1 |
| chr22_+_37678424 | 0.12 |
ENST00000248901.6
|
CYTH4
|
cytohesin 4 |
| chr1_-_176176629 | 0.12 |
ENST00000367669.3
|
RFWD2
|
ring finger and WD repeat domain 2, E3 ubiquitin protein ligase |
| chr4_+_175205038 | 0.12 |
ENST00000457424.2
ENST00000514712.1 |
CEP44
|
centrosomal protein 44kDa |
| chr3_-_129375556 | 0.11 |
ENST00000510323.1
|
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr17_+_55162453 | 0.11 |
ENST00000575322.1
ENST00000337714.3 ENST00000314126.3 |
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr19_+_37837218 | 0.09 |
ENST00000591134.1
|
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chr4_-_156298028 | 0.09 |
ENST00000433024.1
ENST00000379248.2 |
MAP9
|
microtubule-associated protein 9 |
| chr2_+_143886877 | 0.09 |
ENST00000295095.6
|
ARHGAP15
|
Rho GTPase activating protein 15 |
| chr15_+_63889552 | 0.09 |
ENST00000360587.2
|
FBXL22
|
F-box and leucine-rich repeat protein 22 |
| chr8_+_13424352 | 0.09 |
ENST00000297324.4
|
C8orf48
|
chromosome 8 open reading frame 48 |
| chr7_-_86595190 | 0.09 |
ENST00000398276.2
ENST00000416314.1 ENST00000425689.1 |
KIAA1324L
|
KIAA1324-like |
| chr2_+_62900986 | 0.08 |
ENST00000405015.3
ENST00000413434.1 ENST00000426940.1 ENST00000449820.1 |
EHBP1
|
EH domain binding protein 1 |
| chr22_+_45072925 | 0.08 |
ENST00000006251.7
|
PRR5
|
proline rich 5 (renal) |
| chr10_+_71561630 | 0.08 |
ENST00000398974.3
ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr6_-_2842219 | 0.08 |
ENST00000380739.5
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr1_+_89990378 | 0.08 |
ENST00000449440.1
|
LRRC8B
|
leucine rich repeat containing 8 family, member B |
| chr11_+_118398178 | 0.08 |
ENST00000302783.4
ENST00000539546.1 |
TTC36
|
tetratricopeptide repeat domain 36 |
| chr3_-_150264272 | 0.07 |
ENST00000491660.1
ENST00000487153.1 ENST00000239944.2 |
SERP1
|
stress-associated endoplasmic reticulum protein 1 |
| chr10_+_90750493 | 0.07 |
ENST00000357339.2
ENST00000355279.2 |
FAS
|
Fas cell surface death receptor |
| chr3_+_164924716 | 0.07 |
ENST00000470138.1
ENST00000498616.1 |
RP11-85M11.2
|
RP11-85M11.2 |
| chr11_-_119993734 | 0.07 |
ENST00000533302.1
|
TRIM29
|
tripartite motif containing 29 |
| chr19_-_8008533 | 0.07 |
ENST00000597926.1
|
TIMM44
|
translocase of inner mitochondrial membrane 44 homolog (yeast) |
| chr19_-_8675559 | 0.07 |
ENST00000597188.1
|
ADAMTS10
|
ADAM metallopeptidase with thrombospondin type 1 motif, 10 |
| chr16_-_2031464 | 0.06 |
ENST00000356120.4
ENST00000354249.4 |
NOXO1
|
NADPH oxidase organizer 1 |
| chr4_+_69962185 | 0.06 |
ENST00000305231.7
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr14_+_32546274 | 0.06 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr16_-_2770216 | 0.06 |
ENST00000302641.3
|
PRSS27
|
protease, serine 27 |
| chr15_-_90358564 | 0.06 |
ENST00000559874.1
|
ANPEP
|
alanyl (membrane) aminopeptidase |
| chr10_+_17686124 | 0.05 |
ENST00000377524.3
|
STAM
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
| chr12_-_9760482 | 0.05 |
ENST00000229402.3
|
KLRB1
|
killer cell lectin-like receptor subfamily B, member 1 |
| chr12_-_10875831 | 0.05 |
ENST00000279550.7
ENST00000228251.4 |
YBX3
|
Y box binding protein 3 |
| chr15_+_67835005 | 0.05 |
ENST00000178640.5
|
MAP2K5
|
mitogen-activated protein kinase kinase 5 |
| chr8_+_124968225 | 0.05 |
ENST00000399018.1
|
FER1L6
|
fer-1-like 6 (C. elegans) |
| chr15_-_44010458 | 0.04 |
ENST00000541030.1
|
STRC
|
stereocilin |
| chr1_-_145382434 | 0.04 |
ENST00000610154.1
|
RP11-458D21.1
|
RP11-458D21.1 |
| chr11_+_61717816 | 0.04 |
ENST00000435278.2
|
BEST1
|
bestrophin 1 |
| chr10_+_103113802 | 0.04 |
ENST00000370187.3
|
BTRC
|
beta-transducin repeat containing E3 ubiquitin protein ligase |
| chr15_-_65809625 | 0.04 |
ENST00000560436.1
|
DPP8
|
dipeptidyl-peptidase 8 |
| chr2_+_108145913 | 0.04 |
ENST00000443205.1
|
AC096669.3
|
AC096669.3 |
| chr20_+_18548055 | 0.04 |
ENST00000435844.1
ENST00000411646.1 ENST00000608034.1 |
LINC00493
|
long intergenic non-protein coding RNA 493 |
| chr16_-_66583701 | 0.04 |
ENST00000527800.1
ENST00000525974.1 ENST00000563369.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr12_-_54121261 | 0.04 |
ENST00000549784.1
ENST00000262059.4 |
CALCOCO1
|
calcium binding and coiled-coil domain 1 |
| chr10_-_120938303 | 0.04 |
ENST00000356951.3
ENST00000298510.2 |
PRDX3
|
peroxiredoxin 3 |
| chr10_+_106034884 | 0.03 |
ENST00000369707.2
ENST00000429569.2 |
GSTO2
|
glutathione S-transferase omega 2 |
| chr15_-_93616892 | 0.03 |
ENST00000556658.1
ENST00000538818.1 ENST00000425933.2 |
RGMA
|
repulsive guidance molecule family member a |
| chr1_-_146040968 | 0.03 |
ENST00000401010.3
|
NBPF11
|
neuroblastoma breakpoint family, member 11 |
| chr5_-_78281603 | 0.03 |
ENST00000264914.4
|
ARSB
|
arylsulfatase B |
| chr3_+_46204959 | 0.03 |
ENST00000357422.2
|
CCR3
|
chemokine (C-C motif) receptor 3 |
| chr5_-_110848253 | 0.03 |
ENST00000505803.1
ENST00000502322.1 |
STARD4
|
StAR-related lipid transfer (START) domain containing 4 |
| chr17_+_7123125 | 0.02 |
ENST00000356839.5
ENST00000583312.1 ENST00000350303.5 |
ACADVL
|
acyl-CoA dehydrogenase, very long chain |
| chr8_-_94029882 | 0.02 |
ENST00000520686.1
|
TRIQK
|
triple QxxK/R motif containing |
| chr14_-_54420133 | 0.02 |
ENST00000559501.1
ENST00000558984.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr5_-_33984741 | 0.02 |
ENST00000382102.3
ENST00000509381.1 ENST00000342059.3 ENST00000345083.5 |
SLC45A2
|
solute carrier family 45, member 2 |
| chr18_-_47376197 | 0.02 |
ENST00000592688.1
|
MYO5B
|
myosin VB |
| chr9_-_136568822 | 0.02 |
ENST00000371868.1
|
SARDH
|
sarcosine dehydrogenase |
| chr16_+_55522536 | 0.02 |
ENST00000570283.1
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) |
| chr6_-_131299929 | 0.01 |
ENST00000531356.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr17_-_33760269 | 0.01 |
ENST00000452764.3
|
SLFN12
|
schlafen family member 12 |
| chr12_-_49319265 | 0.01 |
ENST00000552878.1
ENST00000453172.2 |
FKBP11
|
FK506 binding protein 11, 19 kDa |
| chr12_+_104680659 | 0.01 |
ENST00000526691.1
ENST00000531691.1 ENST00000388854.3 ENST00000354940.6 ENST00000526390.1 ENST00000531689.1 |
TXNRD1
|
thioredoxin reductase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX19
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0061193 | sensory organ boundary specification(GO:0008052) formation of organ boundary(GO:0010160) taste bud development(GO:0061193) |
| 0.2 | 1.0 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.1 | 0.7 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 0.7 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.1 | 0.7 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.1 | 0.5 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.1 | 0.9 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 0.2 | GO:0061580 | colon epithelial cell migration(GO:0061580) |
| 0.1 | 0.2 | GO:1902573 | positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.1 | 0.6 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.1 | 0.3 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.1 | 0.2 | GO:0010877 | lipid transport involved in lipid storage(GO:0010877) |
| 0.1 | 0.4 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.1 | 0.3 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.1 | 0.7 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.1 | 0.2 | GO:1903450 | regulation of G1 to G0 transition(GO:1903450) positive regulation of G1 to G0 transition(GO:1903452) |
| 0.0 | 0.4 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.0 | 0.9 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.0 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.4 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.3 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.2 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.2 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.2 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 0.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.3 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.2 | GO:0097688 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 | 1.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.3 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.3 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.2 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 1.2 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.6 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.2 | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.3 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.2 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.8 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.4 | GO:0006554 | lysine catabolic process(GO:0006554) |
| 0.0 | 0.6 | GO:0002691 | regulation of cellular extravasation(GO:0002691) |
| 0.0 | 0.2 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.0 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070703 | inner mucus layer(GO:0070702) outer mucus layer(GO:0070703) |
| 0.1 | 0.7 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.1 | 0.4 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.1 | 0.6 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.2 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.3 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.3 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.3 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.7 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.6 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 0.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.2 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 2.4 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 1.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.2 | 0.7 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.2 | 0.6 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.2 | 0.9 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 0.8 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 0.7 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.1 | 0.4 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.1 | 0.7 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.1 | 0.3 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.1 | 0.2 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 0.2 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.3 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.3 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.2 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.0 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 1.2 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 1.8 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.3 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0009374 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.8 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.3 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |