Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
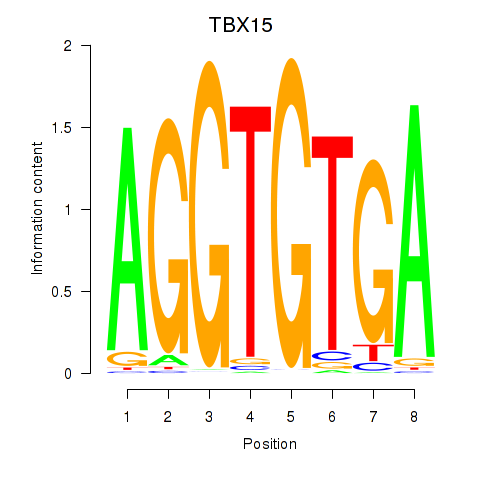
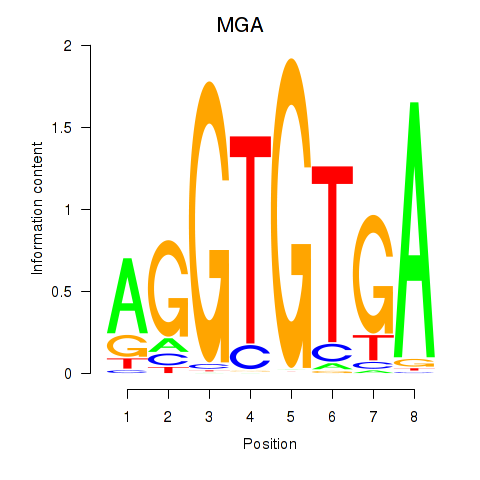
Results for TBX15_MGA
Z-value: 0.19
Transcription factors associated with TBX15_MGA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX15
|
ENSG00000092607.9 | T-box transcription factor 15 |
|
MGA
|
ENSG00000174197.12 | MAX dimerization protein MGA |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX15 | hg19_v2_chr1_-_119532127_119532179 | 0.56 | 4.4e-01 | Click! |
| MGA | hg19_v2_chr15_+_41913690_41913753 | -0.35 | 6.5e-01 | Click! |
Activity profile of TBX15_MGA motif
Sorted Z-values of TBX15_MGA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_46531127 | 0.23 |
ENST00000601033.1
|
CTC-344H19.4
|
CTC-344H19.4 |
| chr5_+_133861339 | 0.13 |
ENST00000282605.4
ENST00000361895.2 ENST00000402835.1 |
PHF15
|
jade family PHD finger 2 |
| chr2_-_27486951 | 0.10 |
ENST00000432351.1
|
SLC30A3
|
solute carrier family 30 (zinc transporter), member 3 |
| chr19_-_46526304 | 0.10 |
ENST00000008938.4
|
PGLYRP1
|
peptidoglycan recognition protein 1 |
| chr8_-_123793048 | 0.09 |
ENST00000607710.1
|
RP11-44N11.2
|
RP11-44N11.2 |
| chrX_+_56259316 | 0.09 |
ENST00000468660.1
|
KLF8
|
Kruppel-like factor 8 |
| chr8_-_25281747 | 0.08 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr11_+_61447845 | 0.07 |
ENST00000257215.5
|
DAGLA
|
diacylglycerol lipase, alpha |
| chr15_-_44092295 | 0.07 |
ENST00000249714.3
ENST00000299969.6 ENST00000319327.6 |
SERINC4
|
serine incorporator 4 |
| chr17_-_39216344 | 0.06 |
ENST00000391418.2
|
KRTAP2-3
|
keratin associated protein 2-3 |
| chr16_-_67965756 | 0.06 |
ENST00000571044.1
ENST00000571605.1 |
CTRL
|
chymotrypsin-like |
| chr2_-_74776586 | 0.06 |
ENST00000420535.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr9_-_34397800 | 0.06 |
ENST00000297623.2
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr3_-_49459865 | 0.06 |
ENST00000427987.1
|
AMT
|
aminomethyltransferase |
| chr12_-_56120838 | 0.06 |
ENST00000548160.1
|
CD63
|
CD63 molecule |
| chr17_-_7232585 | 0.06 |
ENST00000571887.1
ENST00000315614.7 ENST00000399464.2 ENST00000570460.1 |
NEURL4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr22_-_31688431 | 0.06 |
ENST00000402249.3
ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr22_-_31688381 | 0.06 |
ENST00000487265.2
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr6_-_31620149 | 0.06 |
ENST00000435080.1
ENST00000375976.4 ENST00000441054.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr1_-_26633067 | 0.05 |
ENST00000421827.2
ENST00000374215.1 ENST00000374223.1 ENST00000357089.4 ENST00000535108.1 ENST00000314675.7 ENST00000436301.2 ENST00000423664.1 ENST00000374221.3 |
UBXN11
|
UBX domain protein 11 |
| chr12_-_58145889 | 0.05 |
ENST00000547853.1
|
CDK4
|
cyclin-dependent kinase 4 |
| chr2_+_176972000 | 0.05 |
ENST00000249504.5
|
HOXD11
|
homeobox D11 |
| chr12_-_49351228 | 0.05 |
ENST00000541959.1
ENST00000447318.2 |
ARF3
|
ADP-ribosylation factor 3 |
| chr19_-_3500635 | 0.05 |
ENST00000250937.3
|
DOHH
|
deoxyhypusine hydroxylase/monooxygenase |
| chr2_+_201994042 | 0.05 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr1_+_28586006 | 0.05 |
ENST00000253063.3
|
SESN2
|
sestrin 2 |
| chr17_-_34122596 | 0.05 |
ENST00000250144.8
|
MMP28
|
matrix metallopeptidase 28 |
| chr5_-_137514333 | 0.05 |
ENST00000411594.2
ENST00000430331.1 |
BRD8
|
bromodomain containing 8 |
| chr19_-_51875894 | 0.05 |
ENST00000600427.1
ENST00000595217.1 ENST00000221978.5 |
NKG7
|
natural killer cell group 7 sequence |
| chr11_+_122709200 | 0.05 |
ENST00000227348.4
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr3_-_66551351 | 0.05 |
ENST00000273261.3
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr16_+_57139933 | 0.04 |
ENST00000566259.1
|
CPNE2
|
copine II |
| chr17_-_74533734 | 0.04 |
ENST00000589342.1
|
CYGB
|
cytoglobin |
| chr9_+_71986182 | 0.04 |
ENST00000303068.7
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr2_+_29033682 | 0.04 |
ENST00000379579.4
ENST00000334056.5 ENST00000449210.1 |
SPDYA
|
speedy/RINGO cell cycle regulator family member A |
| chr19_+_39279838 | 0.04 |
ENST00000314980.4
|
LGALS7B
|
lectin, galactoside-binding, soluble, 7B |
| chr9_-_86322516 | 0.04 |
ENST00000529923.1
|
UBQLN1
|
ubiquilin 1 |
| chr1_-_44482979 | 0.04 |
ENST00000360584.2
ENST00000357730.2 ENST00000528803.1 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr18_-_59561417 | 0.04 |
ENST00000591306.1
|
RNF152
|
ring finger protein 152 |
| chr19_-_39390212 | 0.04 |
ENST00000437828.1
|
SIRT2
|
sirtuin 2 |
| chr12_+_65672702 | 0.04 |
ENST00000538045.1
ENST00000535239.1 |
MSRB3
|
methionine sulfoxide reductase B3 |
| chr19_-_42916499 | 0.04 |
ENST00000601189.1
ENST00000599211.1 |
LIPE
|
lipase, hormone-sensitive |
| chr17_+_76210367 | 0.04 |
ENST00000592734.1
ENST00000587746.1 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chr22_+_39966758 | 0.04 |
ENST00000407673.1
ENST00000401624.1 ENST00000404898.1 ENST00000402142.3 ENST00000336649.4 ENST00000400164.3 |
CACNA1I
|
calcium channel, voltage-dependent, T type, alpha 1I subunit |
| chr11_+_120081475 | 0.04 |
ENST00000328965.4
|
OAF
|
OAF homolog (Drosophila) |
| chr3_-_49459878 | 0.04 |
ENST00000546031.1
ENST00000458307.2 ENST00000430521.1 |
AMT
|
aminomethyltransferase |
| chr11_-_64889529 | 0.04 |
ENST00000531743.1
ENST00000527548.1 ENST00000526555.1 ENST00000279259.3 |
FAU
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed |
| chr6_+_84563295 | 0.04 |
ENST00000369687.1
|
RIPPLY2
|
ripply transcriptional repressor 2 |
| chrX_+_47050798 | 0.04 |
ENST00000412206.1
ENST00000427561.1 |
UBA1
|
ubiquitin-like modifier activating enzyme 1 |
| chr21_+_30502806 | 0.04 |
ENST00000399928.1
ENST00000399926.1 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr12_+_69742121 | 0.04 |
ENST00000261267.2
ENST00000549690.1 ENST00000548839.1 |
LYZ
|
lysozyme |
| chr3_-_61237050 | 0.04 |
ENST00000476844.1
ENST00000488467.1 ENST00000492590.1 ENST00000468189.1 |
FHIT
|
fragile histidine triad |
| chr9_-_35815013 | 0.04 |
ENST00000259667.5
|
HINT2
|
histidine triad nucleotide binding protein 2 |
| chrX_-_106146547 | 0.04 |
ENST00000276173.4
ENST00000411805.1 |
RIPPLY1
|
ripply transcriptional repressor 1 |
| chr7_-_20826504 | 0.04 |
ENST00000418710.2
ENST00000361443.4 |
SP8
|
Sp8 transcription factor |
| chr13_-_48575443 | 0.04 |
ENST00000378654.3
|
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr11_-_119599794 | 0.04 |
ENST00000264025.3
|
PVRL1
|
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
| chr1_+_8378140 | 0.04 |
ENST00000377479.2
|
SLC45A1
|
solute carrier family 45, member 1 |
| chr11_-_64889649 | 0.04 |
ENST00000434372.2
|
FAU
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed |
| chr7_-_71801980 | 0.04 |
ENST00000329008.5
|
CALN1
|
calneuron 1 |
| chr1_-_205290865 | 0.04 |
ENST00000367157.3
|
NUAK2
|
NUAK family, SNF1-like kinase, 2 |
| chrX_-_153775426 | 0.03 |
ENST00000393562.2
|
G6PD
|
glucose-6-phosphate dehydrogenase |
| chr4_-_73434498 | 0.03 |
ENST00000286657.4
|
ADAMTS3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3 |
| chr12_+_58148842 | 0.03 |
ENST00000266643.5
|
MARCH9
|
membrane-associated ring finger (C3HC4) 9 |
| chr6_-_31619892 | 0.03 |
ENST00000454165.1
ENST00000428326.1 ENST00000452994.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr2_+_219081817 | 0.03 |
ENST00000315717.5
ENST00000420104.1 ENST00000295685.10 |
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr11_-_115127611 | 0.03 |
ENST00000545094.1
|
CADM1
|
cell adhesion molecule 1 |
| chr1_+_15736359 | 0.03 |
ENST00000375980.4
|
EFHD2
|
EF-hand domain family, member D2 |
| chr12_-_56120865 | 0.03 |
ENST00000548898.1
ENST00000552067.1 |
CD63
|
CD63 molecule |
| chr4_-_186732048 | 0.03 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr11_-_85430356 | 0.03 |
ENST00000526999.1
|
SYTL2
|
synaptotagmin-like 2 |
| chr5_+_150040403 | 0.03 |
ENST00000517768.1
ENST00000297130.4 |
MYOZ3
|
myozenin 3 |
| chr11_-_8285405 | 0.03 |
ENST00000335790.3
ENST00000534484.1 |
LMO1
|
LIM domain only 1 (rhombotin 1) |
| chr8_-_145060593 | 0.03 |
ENST00000313059.5
ENST00000524918.1 ENST00000313028.7 ENST00000525773.1 |
PARP10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr5_-_137674000 | 0.03 |
ENST00000510119.1
ENST00000513970.1 |
CDC25C
|
cell division cycle 25C |
| chr6_+_31620191 | 0.03 |
ENST00000375918.2
ENST00000375920.4 |
APOM
|
apolipoprotein M |
| chr9_+_140083099 | 0.03 |
ENST00000322310.5
|
SSNA1
|
Sjogren syndrome nuclear autoantigen 1 |
| chr19_-_10450328 | 0.03 |
ENST00000160262.5
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr11_+_111750206 | 0.03 |
ENST00000530214.1
ENST00000530799.1 |
C11orf1
|
chromosome 11 open reading frame 1 |
| chr7_-_6523688 | 0.03 |
ENST00000490996.1
|
KDELR2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr19_-_10426663 | 0.03 |
ENST00000541276.1
ENST00000393708.3 ENST00000494368.1 |
FDX1L
|
ferredoxin 1-like |
| chrX_+_102862834 | 0.03 |
ENST00000372627.5
ENST00000243286.3 |
TCEAL3
|
transcription elongation factor A (SII)-like 3 |
| chr19_-_7990991 | 0.03 |
ENST00000318978.4
|
CTXN1
|
cortexin 1 |
| chr8_-_11660077 | 0.03 |
ENST00000533405.1
|
RP11-297N6.4
|
Uncharacterized protein |
| chr8_+_56014949 | 0.03 |
ENST00000327381.6
|
XKR4
|
XK, Kell blood group complex subunit-related family, member 4 |
| chr17_-_79869077 | 0.03 |
ENST00000570391.1
|
PCYT2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr6_-_86099898 | 0.03 |
ENST00000455071.1
|
RP11-30P6.6
|
RP11-30P6.6 |
| chr20_-_22565101 | 0.03 |
ENST00000419308.2
|
FOXA2
|
forkhead box A2 |
| chr6_-_36953833 | 0.03 |
ENST00000538808.1
ENST00000460219.1 ENST00000373616.5 ENST00000373627.5 |
MTCH1
|
mitochondrial carrier 1 |
| chr1_-_12677714 | 0.03 |
ENST00000376223.2
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr4_+_102268904 | 0.03 |
ENST00000527564.1
ENST00000529296.1 |
AP001816.1
|
Uncharacterized protein |
| chr7_-_27142290 | 0.03 |
ENST00000222718.5
|
HOXA2
|
homeobox A2 |
| chr6_-_31619742 | 0.03 |
ENST00000433828.1
ENST00000456286.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr7_-_72437484 | 0.03 |
ENST00000395244.1
|
TRIM74
|
tripartite motif containing 74 |
| chr3_+_10857885 | 0.03 |
ENST00000254488.2
ENST00000454147.1 |
SLC6A11
|
solute carrier family 6 (neurotransmitter transporter), member 11 |
| chr2_-_217560248 | 0.03 |
ENST00000233813.4
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr17_-_79869004 | 0.03 |
ENST00000573927.1
ENST00000331285.3 ENST00000572157.1 |
PCYT2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr17_+_77021702 | 0.03 |
ENST00000392445.2
ENST00000354124.3 |
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr3_+_50654821 | 0.03 |
ENST00000457064.1
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr17_-_39203519 | 0.03 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chr20_+_44657845 | 0.03 |
ENST00000243964.3
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5 |
| chr1_-_113247543 | 0.03 |
ENST00000414971.1
ENST00000534717.1 |
RHOC
|
ras homolog family member C |
| chr15_+_40861487 | 0.03 |
ENST00000315616.7
ENST00000559271.1 |
RPUSD2
|
RNA pseudouridylate synthase domain containing 2 |
| chr1_+_145507587 | 0.03 |
ENST00000330165.8
ENST00000369307.3 |
RBM8A
|
RNA binding motif protein 8A |
| chr17_-_40428359 | 0.03 |
ENST00000293328.3
|
STAT5B
|
signal transducer and activator of transcription 5B |
| chr17_-_79869340 | 0.03 |
ENST00000538936.2
|
PCYT2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr11_-_129872672 | 0.03 |
ENST00000531431.1
ENST00000527581.1 |
PRDM10
|
PR domain containing 10 |
| chr22_-_42342733 | 0.03 |
ENST00000402420.1
|
CENPM
|
centromere protein M |
| chr11_-_66112555 | 0.03 |
ENST00000425825.2
ENST00000359957.3 |
BRMS1
|
breast cancer metastasis suppressor 1 |
| chr3_+_107364683 | 0.03 |
ENST00000413213.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chr17_-_1553346 | 0.03 |
ENST00000301336.6
|
RILP
|
Rab interacting lysosomal protein |
| chr7_+_107220899 | 0.03 |
ENST00000379117.2
ENST00000473124.1 |
BCAP29
|
B-cell receptor-associated protein 29 |
| chr17_-_79869243 | 0.03 |
ENST00000538721.2
ENST00000573636.2 ENST00000571105.1 ENST00000576343.1 ENST00000572473.1 |
PCYT2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr11_-_76155618 | 0.03 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr3_-_120169828 | 0.03 |
ENST00000424703.2
ENST00000469005.1 |
FSTL1
|
follistatin-like 1 |
| chr8_+_22019168 | 0.03 |
ENST00000318561.3
ENST00000521315.1 ENST00000437090.2 ENST00000520605.1 ENST00000522109.1 ENST00000524255.1 ENST00000523296.1 ENST00000518615.1 |
SFTPC
|
surfactant protein C |
| chr11_+_46402297 | 0.03 |
ENST00000405308.2
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr17_+_25958174 | 0.03 |
ENST00000313648.6
ENST00000577392.1 ENST00000584661.1 ENST00000413914.2 |
LGALS9
|
lectin, galactoside-binding, soluble, 9 |
| chr3_+_107364769 | 0.03 |
ENST00000449271.1
ENST00000425868.1 ENST00000449213.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr17_+_46184911 | 0.03 |
ENST00000580219.1
ENST00000452859.2 ENST00000393405.2 ENST00000439357.2 ENST00000359238.2 |
SNX11
|
sorting nexin 11 |
| chr12_+_53400176 | 0.03 |
ENST00000551002.1
ENST00000420463.3 ENST00000416762.3 ENST00000549481.1 ENST00000552490.1 |
EIF4B
|
eukaryotic translation initiation factor 4B |
| chr11_+_64889773 | 0.03 |
ENST00000534078.1
ENST00000526171.1 ENST00000279242.2 ENST00000531705.1 ENST00000533943.1 |
MRPL49
|
mitochondrial ribosomal protein L49 |
| chr19_-_41220540 | 0.03 |
ENST00000594490.1
|
ADCK4
|
aarF domain containing kinase 4 |
| chr11_-_119993979 | 0.02 |
ENST00000524816.3
ENST00000525327.1 |
TRIM29
|
tripartite motif containing 29 |
| chr16_-_69418553 | 0.02 |
ENST00000569542.2
|
TERF2
|
telomeric repeat binding factor 2 |
| chr12_-_56121612 | 0.02 |
ENST00000546939.1
|
CD63
|
CD63 molecule |
| chr4_-_146859623 | 0.02 |
ENST00000379448.4
ENST00000513320.1 |
ZNF827
|
zinc finger protein 827 |
| chr8_-_27457494 | 0.02 |
ENST00000521770.1
|
CLU
|
clusterin |
| chr8_+_1993173 | 0.02 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr2_-_42160486 | 0.02 |
ENST00000427054.1
|
AC104654.2
|
AC104654.2 |
| chr19_-_42721819 | 0.02 |
ENST00000336034.4
ENST00000598200.1 ENST00000598727.1 ENST00000596251.1 |
DEDD2
|
death effector domain containing 2 |
| chr8_+_22435762 | 0.02 |
ENST00000456545.1
|
PDLIM2
|
PDZ and LIM domain 2 (mystique) |
| chr21_-_46962379 | 0.02 |
ENST00000311124.4
ENST00000380010.4 |
SLC19A1
|
solute carrier family 19 (folate transporter), member 1 |
| chr1_+_207002222 | 0.02 |
ENST00000270218.6
|
IL19
|
interleukin 19 |
| chr4_-_184241927 | 0.02 |
ENST00000323319.5
|
CLDN22
|
claudin 22 |
| chr16_-_28550320 | 0.02 |
ENST00000395641.2
|
NUPR1
|
nuclear protein, transcriptional regulator, 1 |
| chr12_+_57853918 | 0.02 |
ENST00000532291.1
ENST00000543426.1 ENST00000228682.2 ENST00000546141.1 |
GLI1
|
GLI family zinc finger 1 |
| chr3_-_125094093 | 0.02 |
ENST00000484491.1
ENST00000492394.1 ENST00000471196.1 ENST00000468369.1 ENST00000544464.1 ENST00000485866.1 ENST00000360647.4 |
ZNF148
|
zinc finger protein 148 |
| chr2_+_172864490 | 0.02 |
ENST00000315796.4
|
METAP1D
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr7_+_99036996 | 0.02 |
ENST00000441580.1
ENST00000412686.1 |
CPSF4
|
cleavage and polyadenylation specific factor 4, 30kDa |
| chr7_+_76090993 | 0.02 |
ENST00000425780.1
ENST00000456590.1 ENST00000451769.1 ENST00000324432.5 ENST00000307569.8 ENST00000457529.1 ENST00000446600.1 ENST00000413936.2 ENST00000423646.1 ENST00000438930.1 ENST00000430490.2 |
DTX2
|
deltex homolog 2 (Drosophila) |
| chr6_-_107436473 | 0.02 |
ENST00000369042.1
|
BEND3
|
BEN domain containing 3 |
| chr16_-_4395905 | 0.02 |
ENST00000571941.1
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr18_+_73118967 | 0.02 |
ENST00000578340.1
|
RP11-321M21.3
|
HCG1996301; Uncharacterized protein |
| chr3_+_48264816 | 0.02 |
ENST00000296435.2
ENST00000576243.1 |
CAMP
|
cathelicidin antimicrobial peptide |
| chrX_-_53461288 | 0.02 |
ENST00000375298.4
ENST00000375304.5 |
HSD17B10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr5_+_137514687 | 0.02 |
ENST00000394894.3
|
KIF20A
|
kinesin family member 20A |
| chr7_+_34697846 | 0.02 |
ENST00000381553.3
ENST00000465305.1 ENST00000360581.1 ENST00000381542.1 ENST00000359791.1 ENST00000531252.1 |
NPSR1
|
neuropeptide S receptor 1 |
| chr6_-_160166218 | 0.02 |
ENST00000537657.1
|
SOD2
|
superoxide dismutase 2, mitochondrial |
| chrX_+_54556633 | 0.02 |
ENST00000336470.4
ENST00000360845.2 |
GNL3L
|
guanine nucleotide binding protein-like 3 (nucleolar)-like |
| chr6_-_30815936 | 0.02 |
ENST00000442852.1
|
XXbac-BPG27H4.8
|
XXbac-BPG27H4.8 |
| chr20_+_1115821 | 0.02 |
ENST00000435720.1
|
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
| chr7_-_6066183 | 0.02 |
ENST00000422786.1
|
EIF2AK1
|
eukaryotic translation initiation factor 2-alpha kinase 1 |
| chr11_+_65687158 | 0.02 |
ENST00000532933.1
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr11_-_64703354 | 0.02 |
ENST00000532246.1
ENST00000279168.2 |
GPHA2
|
glycoprotein hormone alpha 2 |
| chr16_-_66968055 | 0.02 |
ENST00000568572.1
|
FAM96B
|
family with sequence similarity 96, member B |
| chr17_-_34890037 | 0.02 |
ENST00000589404.1
|
MYO19
|
myosin XIX |
| chr8_+_22462532 | 0.02 |
ENST00000389279.3
|
CCAR2
|
cell cycle and apoptosis regulator 2 |
| chrX_+_48660107 | 0.02 |
ENST00000376643.2
|
HDAC6
|
histone deacetylase 6 |
| chr17_+_34431212 | 0.02 |
ENST00000394495.1
|
CCL4
|
chemokine (C-C motif) ligand 4 |
| chr3_-_66551397 | 0.02 |
ENST00000383703.3
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr7_+_141438393 | 0.02 |
ENST00000484178.1
ENST00000473783.1 ENST00000481508.1 |
SSBP1
|
single-stranded DNA binding protein 1, mitochondrial |
| chr11_+_64950801 | 0.02 |
ENST00000526468.1
|
CAPN1
|
calpain 1, (mu/I) large subunit |
| chr9_-_35812140 | 0.02 |
ENST00000497810.1
ENST00000396638.2 ENST00000484764.1 |
SPAG8
|
sperm associated antigen 8 |
| chr12_-_49351303 | 0.02 |
ENST00000256682.4
|
ARF3
|
ADP-ribosylation factor 3 |
| chr18_+_32073253 | 0.02 |
ENST00000283365.9
ENST00000596745.1 ENST00000315456.6 |
DTNA
|
dystrobrevin, alpha |
| chr8_-_144679602 | 0.02 |
ENST00000526710.1
|
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr4_-_71532601 | 0.02 |
ENST00000510614.1
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr8_+_28748765 | 0.02 |
ENST00000355231.5
|
HMBOX1
|
homeobox containing 1 |
| chr16_-_67260901 | 0.02 |
ENST00000341546.3
ENST00000409509.1 ENST00000433915.1 ENST00000454102.2 |
LRRC29
AC040160.1
|
leucine rich repeat containing 29 Uncharacterized protein; cDNA FLJ57407, weakly similar to Mus musculus leucine rich repeat containing 29 (Lrrc29), mRNA |
| chr4_-_71532668 | 0.02 |
ENST00000510437.1
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr4_+_144354644 | 0.02 |
ENST00000512843.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr12_-_56121580 | 0.02 |
ENST00000550776.1
|
CD63
|
CD63 molecule |
| chr16_+_30662360 | 0.02 |
ENST00000542965.2
|
PRR14
|
proline rich 14 |
| chr7_+_99156393 | 0.02 |
ENST00000422164.1
ENST00000422647.1 ENST00000427931.1 |
ZNF655
|
zinc finger protein 655 |
| chr17_+_34430980 | 0.02 |
ENST00000250151.4
|
CCL4
|
chemokine (C-C motif) ligand 4 |
| chrX_-_48693955 | 0.02 |
ENST00000218230.5
|
PCSK1N
|
proprotein convertase subtilisin/kexin type 1 inhibitor |
| chr17_+_7590734 | 0.02 |
ENST00000457584.2
|
WRAP53
|
WD repeat containing, antisense to TP53 |
| chr17_-_79869228 | 0.02 |
ENST00000570388.1
|
PCYT2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr11_-_47616210 | 0.02 |
ENST00000302514.3
|
C1QTNF4
|
C1q and tumor necrosis factor related protein 4 |
| chr6_+_36646435 | 0.02 |
ENST00000244741.5
ENST00000405375.1 ENST00000373711.2 |
CDKN1A
|
cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
| chr3_-_120170052 | 0.02 |
ENST00000295633.3
|
FSTL1
|
follistatin-like 1 |
| chr2_-_74648702 | 0.02 |
ENST00000518863.1
|
C2orf81
|
chromosome 2 open reading frame 81 |
| chr16_+_67906919 | 0.02 |
ENST00000358933.5
|
EDC4
|
enhancer of mRNA decapping 4 |
| chr11_+_46402482 | 0.02 |
ENST00000441869.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr1_+_44444865 | 0.02 |
ENST00000372324.1
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr19_+_17337473 | 0.02 |
ENST00000598068.1
|
OCEL1
|
occludin/ELL domain containing 1 |
| chrX_-_128788914 | 0.02 |
ENST00000429967.1
ENST00000307484.6 |
APLN
|
apelin |
| chr22_-_20231207 | 0.02 |
ENST00000425986.1
|
RTN4R
|
reticulon 4 receptor |
| chr10_-_131762105 | 0.02 |
ENST00000368648.3
ENST00000355311.5 |
EBF3
|
early B-cell factor 3 |
| chr1_+_41445413 | 0.02 |
ENST00000541520.1
|
CTPS1
|
CTP synthase 1 |
| chrX_+_99899180 | 0.02 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr6_+_30294612 | 0.02 |
ENST00000440271.1
ENST00000396551.3 ENST00000376656.4 ENST00000540416.1 ENST00000428728.1 ENST00000396548.1 ENST00000428404.1 |
TRIM39
|
tripartite motif containing 39 |
| chr14_+_95078714 | 0.02 |
ENST00000393078.3
ENST00000393080.4 ENST00000467132.1 |
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chrX_-_132549506 | 0.02 |
ENST00000370828.3
|
GPC4
|
glypican 4 |
| chr9_-_96717654 | 0.02 |
ENST00000253968.6
|
BARX1
|
BARX homeobox 1 |
| chr21_+_37507210 | 0.02 |
ENST00000290354.5
|
CBR3
|
carbonyl reductase 3 |
| chr5_+_172332220 | 0.02 |
ENST00000518247.1
ENST00000326654.2 |
ERGIC1
|
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
| chr16_-_67260691 | 0.02 |
ENST00000447579.1
ENST00000393992.1 ENST00000424285.1 |
LRRC29
|
leucine rich repeat containing 29 |
| chr19_-_10450287 | 0.02 |
ENST00000589261.1
ENST00000590569.1 ENST00000589580.1 ENST00000589249.1 |
ICAM3
|
intercellular adhesion molecule 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX15_MGA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032826 | natural killer cell differentiation involved in immune response(GO:0002325) regulation of natural killer cell differentiation involved in immune response(GO:0032826) regulation of cytolysis in other organism(GO:0051710) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.0 | GO:1901340 | negative regulation of store-operated calcium channel activity(GO:1901340) |
| 0.0 | 0.0 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.0 | GO:0061433 | cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 0.0 | 0.0 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.0 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.1 | GO:2000329 | peptidyl-lysine oxidation(GO:0018057) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.0 | 0.1 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.0 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.0 | 0.0 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.1 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.0 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.1 | GO:0015633 | zinc transporting ATPase activity(GO:0015633) |
| 0.0 | 0.1 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.0 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.0 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.0 | GO:0046970 | NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.0 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.0 | 0.0 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.0 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |