Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
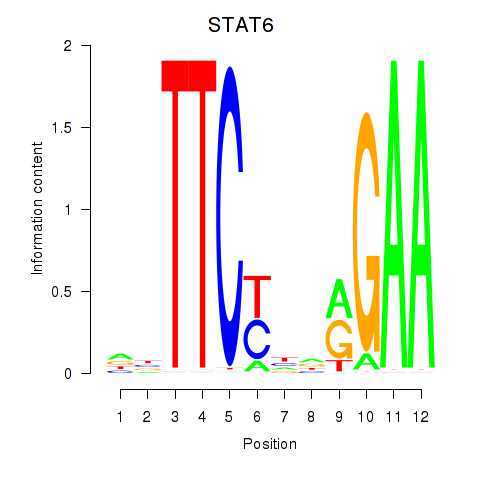
Results for STAT6
Z-value: 0.63
Transcription factors associated with STAT6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
STAT6
|
ENSG00000166888.6 | signal transducer and activator of transcription 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| STAT6 | hg19_v2_chr12_-_57504069_57504123 | -0.91 | 9.2e-02 | Click! |
Activity profile of STAT6 motif
Sorted Z-values of STAT6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_3469181 | 0.78 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr5_+_667759 | 0.63 |
ENST00000594226.1
|
AC026740.1
|
Uncharacterized protein |
| chr1_-_179834311 | 0.51 |
ENST00000553856.1
|
IFRG15
|
Homo sapiens torsin A interacting protein 2 (TOR1AIP2), transcript variant 1, mRNA. |
| chr7_-_50628745 | 0.48 |
ENST00000380984.4
ENST00000357936.5 ENST00000426377.1 |
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase) |
| chr3_+_172034218 | 0.47 |
ENST00000366261.2
|
AC092964.1
|
Uncharacterized protein |
| chr15_+_96869165 | 0.44 |
ENST00000421109.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr1_-_36020531 | 0.41 |
ENST00000440579.1
ENST00000494948.1 |
KIAA0319L
|
KIAA0319-like |
| chr4_-_120988229 | 0.38 |
ENST00000296509.6
|
MAD2L1
|
MAD2 mitotic arrest deficient-like 1 (yeast) |
| chr1_+_45965725 | 0.38 |
ENST00000401061.4
|
MMACHC
|
methylmalonic aciduria (cobalamin deficiency) cblC type, with homocystinuria |
| chr7_+_106415457 | 0.36 |
ENST00000490162.2
ENST00000470135.1 |
RP5-884M6.1
|
RP5-884M6.1 |
| chr21_+_17791648 | 0.35 |
ENST00000602892.1
ENST00000418813.2 ENST00000435697.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr11_-_60720002 | 0.33 |
ENST00000538739.1
|
SLC15A3
|
solute carrier family 15 (oligopeptide transporter), member 3 |
| chr16_+_57728701 | 0.33 |
ENST00000569375.1
ENST00000360716.3 ENST00000569167.1 ENST00000394337.4 ENST00000563126.1 ENST00000336825.8 |
CCDC135
|
coiled-coil domain containing 135 |
| chr11_+_76745385 | 0.33 |
ENST00000533140.1
ENST00000354301.5 ENST00000528622.1 |
B3GNT6
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 6 (core 3 synthase) |
| chr2_-_204400113 | 0.32 |
ENST00000319170.5
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr1_+_61869748 | 0.31 |
ENST00000357977.5
|
NFIA
|
nuclear factor I/A |
| chr6_+_31730773 | 0.30 |
ENST00000415669.2
ENST00000425424.1 |
SAPCD1
|
suppressor APC domain containing 1 |
| chr2_-_120124258 | 0.30 |
ENST00000409877.1
ENST00000409523.1 ENST00000409466.2 ENST00000414534.1 |
C2orf76
|
chromosome 2 open reading frame 76 |
| chr5_+_110427983 | 0.29 |
ENST00000513710.2
ENST00000505303.1 |
WDR36
|
WD repeat domain 36 |
| chr6_+_31540056 | 0.29 |
ENST00000418386.2
|
LTA
|
lymphotoxin alpha |
| chr2_-_204400013 | 0.29 |
ENST00000374489.2
ENST00000374488.2 ENST00000308091.4 ENST00000453034.1 ENST00000420371.1 |
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr4_-_110723134 | 0.28 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr9_-_100000957 | 0.28 |
ENST00000366109.2
ENST00000607322.1 |
RP11-498P14.5
|
RP11-498P14.5 |
| chr6_+_30525051 | 0.27 |
ENST00000376557.3
|
PRR3
|
proline rich 3 |
| chr7_-_38969150 | 0.27 |
ENST00000418457.2
|
VPS41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr1_+_42619070 | 0.26 |
ENST00000372581.1
|
GUCA2B
|
guanylate cyclase activator 2B (uroguanylin) |
| chr8_+_97506033 | 0.26 |
ENST00000518385.1
|
SDC2
|
syndecan 2 |
| chr2_-_120124383 | 0.26 |
ENST00000334816.7
|
C2orf76
|
chromosome 2 open reading frame 76 |
| chr1_+_45274154 | 0.25 |
ENST00000450269.1
ENST00000453418.1 ENST00000409335.2 |
BTBD19
|
BTB (POZ) domain containing 19 |
| chr2_+_145425573 | 0.25 |
ENST00000600064.1
ENST00000597670.1 ENST00000414256.1 ENST00000599187.1 ENST00000451774.1 ENST00000599072.1 ENST00000596589.1 ENST00000597893.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr3_+_63898275 | 0.24 |
ENST00000538065.1
|
ATXN7
|
ataxin 7 |
| chr5_-_151066514 | 0.24 |
ENST00000538026.1
ENST00000522348.1 ENST00000521569.1 |
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr18_+_66382428 | 0.23 |
ENST00000578970.1
ENST00000582371.1 ENST00000584775.1 |
CCDC102B
|
coiled-coil domain containing 102B |
| chr17_+_68071389 | 0.23 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr11_+_73358594 | 0.22 |
ENST00000227214.6
ENST00000398494.4 ENST00000543085.1 |
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr1_-_241803679 | 0.22 |
ENST00000331838.5
|
OPN3
|
opsin 3 |
| chr3_-_57233966 | 0.22 |
ENST00000473921.1
ENST00000295934.3 |
HESX1
|
HESX homeobox 1 |
| chr11_-_67981046 | 0.22 |
ENST00000402789.1
ENST00000402185.2 ENST00000458496.1 |
SUV420H1
|
suppressor of variegation 4-20 homolog 1 (Drosophila) |
| chr3_+_153839149 | 0.22 |
ENST00000465093.1
ENST00000465817.1 |
ARHGEF26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chrX_-_106960285 | 0.21 |
ENST00000503515.1
ENST00000372397.2 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr14_-_37641618 | 0.21 |
ENST00000555449.1
|
SLC25A21
|
solute carrier family 25 (mitochondrial oxoadipate carrier), member 21 |
| chr2_+_145425534 | 0.20 |
ENST00000432608.1
ENST00000597655.1 ENST00000598659.1 ENST00000600679.1 ENST00000601277.1 ENST00000451027.1 ENST00000445791.1 ENST00000596540.1 ENST00000596230.1 ENST00000594471.1 ENST00000598248.1 ENST00000597469.1 ENST00000431734.1 ENST00000595686.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr10_+_115674530 | 0.20 |
ENST00000451472.1
|
AL162407.1
|
CDNA FLJ20147 fis, clone COL07954; HCG1781466; Uncharacterized protein |
| chr3_-_45957088 | 0.20 |
ENST00000539217.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr1_-_241799232 | 0.19 |
ENST00000366553.1
|
CHML
|
choroideremia-like (Rab escort protein 2) |
| chr17_-_60005365 | 0.19 |
ENST00000444766.3
|
INTS2
|
integrator complex subunit 2 |
| chr2_-_203736150 | 0.18 |
ENST00000457524.1
ENST00000421334.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr15_+_69857515 | 0.17 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr1_-_68299130 | 0.17 |
ENST00000370982.3
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12 |
| chr16_+_70207686 | 0.17 |
ENST00000541793.2
ENST00000314151.8 ENST00000565806.1 ENST00000569347.2 ENST00000536907.2 |
CLEC18C
|
C-type lectin domain family 18, member C |
| chr4_+_95972822 | 0.17 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr19_+_13228917 | 0.17 |
ENST00000586171.1
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr16_-_74455290 | 0.16 |
ENST00000339953.5
|
CLEC18B
|
C-type lectin domain family 18, member B |
| chr11_+_36616044 | 0.16 |
ENST00000334307.5
ENST00000531554.1 ENST00000347206.4 ENST00000534635.1 ENST00000446510.2 ENST00000530697.1 ENST00000527108.1 |
C11orf74
|
chromosome 11 open reading frame 74 |
| chr2_-_230786378 | 0.16 |
ENST00000430954.1
|
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr17_-_34890617 | 0.15 |
ENST00000586886.1
ENST00000585719.1 ENST00000585818.1 |
MYO19
|
myosin XIX |
| chr1_+_150245099 | 0.15 |
ENST00000369099.3
|
C1orf54
|
chromosome 1 open reading frame 54 |
| chr1_+_150245177 | 0.15 |
ENST00000369098.3
|
C1orf54
|
chromosome 1 open reading frame 54 |
| chr17_+_34538310 | 0.15 |
ENST00000444414.1
ENST00000378350.4 ENST00000389068.5 ENST00000588929.1 ENST00000589079.1 ENST00000589336.1 ENST00000400702.4 ENST00000591167.1 ENST00000586598.1 ENST00000591637.1 ENST00000378352.4 ENST00000358756.5 |
CCL4L1
|
chemokine (C-C motif) ligand 4-like 1 |
| chr7_+_112120908 | 0.15 |
ENST00000439068.2
ENST00000312849.4 ENST00000429049.1 |
LSMEM1
|
leucine-rich single-pass membrane protein 1 |
| chr2_-_230786032 | 0.14 |
ENST00000428959.1
|
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr20_-_18477862 | 0.14 |
ENST00000337227.4
|
RBBP9
|
retinoblastoma binding protein 9 |
| chr6_+_147527103 | 0.14 |
ENST00000179882.6
|
STXBP5
|
syntaxin binding protein 5 (tomosyn) |
| chr2_+_219745020 | 0.14 |
ENST00000258411.3
|
WNT10A
|
wingless-type MMTV integration site family, member 10A |
| chrX_-_72434628 | 0.14 |
ENST00000536638.1
ENST00000373517.3 |
NAP1L2
|
nucleosome assembly protein 1-like 2 |
| chr16_-_29757272 | 0.13 |
ENST00000329410.3
|
C16orf54
|
chromosome 16 open reading frame 54 |
| chr2_+_202316392 | 0.13 |
ENST00000194530.3
ENST00000392249.2 |
STRADB
|
STE20-related kinase adaptor beta |
| chr3_-_120365866 | 0.13 |
ENST00000475447.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr4_+_141677577 | 0.13 |
ENST00000609937.1
|
RP11-102N12.3
|
RP11-102N12.3 |
| chr11_+_118230287 | 0.13 |
ENST00000252108.3
ENST00000431736.2 |
UBE4A
|
ubiquitination factor E4A |
| chr21_+_17791838 | 0.13 |
ENST00000453910.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr10_-_61122220 | 0.12 |
ENST00000422313.2
ENST00000435852.2 ENST00000442566.3 ENST00000373868.2 ENST00000277705.6 ENST00000373867.3 ENST00000419214.2 |
FAM13C
|
family with sequence similarity 13, member C |
| chr6_+_26440700 | 0.12 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr1_+_26759295 | 0.12 |
ENST00000430232.1
|
DHDDS
|
dehydrodolichyl diphosphate synthase |
| chr13_+_73629107 | 0.12 |
ENST00000539231.1
|
KLF5
|
Kruppel-like factor 5 (intestinal) |
| chr15_+_101402041 | 0.12 |
ENST00000558475.1
ENST00000558641.1 ENST00000559673.1 |
RP11-66B24.1
|
RP11-66B24.1 |
| chr3_-_45957534 | 0.12 |
ENST00000536047.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr2_-_190044480 | 0.11 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chrX_-_138914394 | 0.11 |
ENST00000327569.3
ENST00000361648.2 ENST00000370543.1 ENST00000359686.2 |
ATP11C
|
ATPase, class VI, type 11C |
| chrX_-_73511908 | 0.11 |
ENST00000455395.1
|
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chr3_-_37218023 | 0.11 |
ENST00000416425.1
|
LRRFIP2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr8_+_104311059 | 0.11 |
ENST00000358755.4
ENST00000523739.1 ENST00000540287.1 |
FZD6
|
frizzled family receptor 6 |
| chr15_+_85525205 | 0.10 |
ENST00000394553.1
ENST00000339708.5 |
PDE8A
|
phosphodiesterase 8A |
| chr8_-_117886732 | 0.10 |
ENST00000517485.1
|
RAD21
|
RAD21 homolog (S. pombe) |
| chr14_-_24732738 | 0.10 |
ENST00000558074.1
ENST00000560226.1 |
TGM1
|
transglutaminase 1 |
| chr9_+_100000717 | 0.10 |
ENST00000375205.2
ENST00000357054.1 ENST00000395220.1 ENST00000375202.2 ENST00000411667.2 |
CCDC180
|
coiled-coil domain containing 180 |
| chr2_+_7118755 | 0.10 |
ENST00000433456.1
|
RNF144A
|
ring finger protein 144A |
| chr17_+_7384721 | 0.10 |
ENST00000412468.2
|
SLC35G6
|
solute carrier family 35, member G6 |
| chr5_+_118691706 | 0.09 |
ENST00000415806.2
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr14_-_23285011 | 0.09 |
ENST00000397532.3
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr7_+_23637763 | 0.09 |
ENST00000410069.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chr9_-_131486367 | 0.09 |
ENST00000372663.4
ENST00000406904.2 ENST00000452105.1 ENST00000372672.2 ENST00000372667.5 |
ZDHHC12
|
zinc finger, DHHC-type containing 12 |
| chr4_+_110769258 | 0.09 |
ENST00000594814.1
|
LRIT3
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3 |
| chr15_+_30918879 | 0.09 |
ENST00000428041.2
|
ARHGAP11B
|
Rho GTPase activating protein 11B |
| chr14_-_23285069 | 0.09 |
ENST00000554758.1
ENST00000397528.4 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr8_+_55466915 | 0.09 |
ENST00000522711.2
|
RP11-53M11.3
|
RP11-53M11.3 |
| chr10_-_35379524 | 0.09 |
ENST00000374751.3
ENST00000374742.1 ENST00000602371.1 |
CUL2
|
cullin 2 |
| chr3_+_189507432 | 0.09 |
ENST00000354600.5
|
TP63
|
tumor protein p63 |
| chr11_-_14379997 | 0.09 |
ENST00000526063.1
ENST00000532814.1 |
RRAS2
|
related RAS viral (r-ras) oncogene homolog 2 |
| chr20_-_55100981 | 0.09 |
ENST00000243913.4
|
GCNT7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr12_+_12764773 | 0.08 |
ENST00000228865.2
|
CREBL2
|
cAMP responsive element binding protein-like 2 |
| chr4_+_166300084 | 0.08 |
ENST00000402744.4
|
CPE
|
carboxypeptidase E |
| chr8_-_117886955 | 0.08 |
ENST00000297338.2
|
RAD21
|
RAD21 homolog (S. pombe) |
| chr10_-_35379241 | 0.08 |
ENST00000374748.1
ENST00000374749.3 |
CUL2
|
cullin 2 |
| chr8_-_37594944 | 0.08 |
ENST00000330539.1
|
RP11-863K10.7
|
Uncharacterized protein |
| chr21_+_17792672 | 0.08 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr13_-_45915221 | 0.08 |
ENST00000309246.5
ENST00000379060.4 ENST00000379055.1 ENST00000527226.1 ENST00000379056.1 |
TPT1
|
tumor protein, translationally-controlled 1 |
| chr2_-_204399976 | 0.08 |
ENST00000457812.1
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr2_-_39664206 | 0.08 |
ENST00000484274.1
|
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr6_-_24877490 | 0.08 |
ENST00000540914.1
ENST00000378023.4 |
FAM65B
|
family with sequence similarity 65, member B |
| chr10_+_75541796 | 0.08 |
ENST00000372837.3
ENST00000372833.5 |
CHCHD1
|
coiled-coil-helix-coiled-coil-helix domain containing 1 |
| chr15_-_64386120 | 0.08 |
ENST00000300030.3
|
FAM96A
|
family with sequence similarity 96, member A |
| chr17_+_34639793 | 0.07 |
ENST00000394465.2
ENST00000394463.2 ENST00000378342.4 |
CCL4L2
|
chemokine (C-C motif) ligand 4-like 2 |
| chr9_-_13175823 | 0.07 |
ENST00000545857.1
|
MPDZ
|
multiple PDZ domain protein |
| chr2_+_27760247 | 0.07 |
ENST00000447166.1
|
AC109829.1
|
Uncharacterized protein |
| chr5_+_138609441 | 0.07 |
ENST00000509990.1
ENST00000506147.1 ENST00000512107.1 |
MATR3
|
matrin 3 |
| chr12_+_6493319 | 0.07 |
ENST00000536876.1
|
LTBR
|
lymphotoxin beta receptor (TNFR superfamily, member 3) |
| chr12_+_44152740 | 0.07 |
ENST00000440781.2
ENST00000431837.1 ENST00000550616.1 ENST00000448290.2 ENST00000551736.1 |
IRAK4
|
interleukin-1 receptor-associated kinase 4 |
| chr5_+_35856951 | 0.07 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr18_-_67615088 | 0.07 |
ENST00000582621.1
|
CD226
|
CD226 molecule |
| chr6_+_84222220 | 0.06 |
ENST00000369700.3
|
PRSS35
|
protease, serine, 35 |
| chr11_-_67980744 | 0.06 |
ENST00000401547.2
ENST00000453170.1 ENST00000304363.4 |
SUV420H1
|
suppressor of variegation 4-20 homolog 1 (Drosophila) |
| chr22_+_32439019 | 0.06 |
ENST00000266088.4
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr3_+_189507460 | 0.06 |
ENST00000434928.1
|
TP63
|
tumor protein p63 |
| chr1_-_227505826 | 0.06 |
ENST00000334218.5
ENST00000366766.2 ENST00000366764.2 |
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like) |
| chr3_+_179370517 | 0.06 |
ENST00000263966.3
|
USP13
|
ubiquitin specific peptidase 13 (isopeptidase T-3) |
| chr2_-_202316260 | 0.06 |
ENST00000332624.3
|
TRAK2
|
trafficking protein, kinesin binding 2 |
| chr16_+_69985083 | 0.06 |
ENST00000288040.6
ENST00000449317.2 |
CLEC18A
|
C-type lectin domain family 18, member A |
| chr9_+_36572851 | 0.05 |
ENST00000298048.2
ENST00000538311.1 ENST00000536987.1 ENST00000545008.1 ENST00000536860.1 ENST00000536329.1 ENST00000541717.1 ENST00000543751.1 |
MELK
|
maternal embryonic leucine zipper kinase |
| chr14_-_24551137 | 0.05 |
ENST00000396995.1
|
NRL
|
neural retina leucine zipper |
| chrX_+_100878112 | 0.05 |
ENST00000491568.2
ENST00000479298.1 |
ARMCX3
|
armadillo repeat containing, X-linked 3 |
| chr15_+_76030311 | 0.05 |
ENST00000543887.1
|
AC019294.1
|
AC019294.1 |
| chr18_-_21017817 | 0.05 |
ENST00000542162.1
ENST00000383233.3 ENST00000582336.1 ENST00000450466.2 ENST00000578520.1 ENST00000399707.1 |
TMEM241
|
transmembrane protein 241 |
| chr14_-_37642016 | 0.05 |
ENST00000331299.5
|
SLC25A21
|
solute carrier family 25 (mitochondrial oxoadipate carrier), member 21 |
| chr11_-_10828892 | 0.05 |
ENST00000525681.1
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr2_-_203735976 | 0.05 |
ENST00000435143.1
|
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr2_-_134326009 | 0.04 |
ENST00000409261.1
ENST00000409213.1 |
NCKAP5
|
NCK-associated protein 5 |
| chr6_-_11232891 | 0.04 |
ENST00000379433.5
ENST00000379446.5 |
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr12_-_109747023 | 0.04 |
ENST00000355216.1
ENST00000299162.5 |
FOXN4
|
forkhead box N4 |
| chr16_+_82090028 | 0.04 |
ENST00000568090.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr20_+_57209828 | 0.04 |
ENST00000598340.1
|
MGC4294
|
HCG1785561; MGC4294 protein; Uncharacterized protein |
| chr5_-_74162739 | 0.04 |
ENST00000513277.1
|
FAM169A
|
family with sequence similarity 169, member A |
| chr3_+_51575596 | 0.04 |
ENST00000409535.2
|
RAD54L2
|
RAD54-like 2 (S. cerevisiae) |
| chr10_+_98064085 | 0.04 |
ENST00000419175.1
ENST00000371174.2 |
DNTT
|
DNA nucleotidylexotransferase |
| chr3_+_141121164 | 0.04 |
ENST00000510338.1
ENST00000504673.1 |
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr1_+_202431859 | 0.04 |
ENST00000391959.3
ENST00000367270.4 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr12_+_26164645 | 0.04 |
ENST00000542004.1
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr1_-_108735440 | 0.04 |
ENST00000370041.4
|
SLC25A24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr2_+_182756915 | 0.03 |
ENST00000428267.2
|
SSFA2
|
sperm specific antigen 2 |
| chr17_-_62308087 | 0.03 |
ENST00000583097.1
|
TEX2
|
testis expressed 2 |
| chr1_-_153321301 | 0.03 |
ENST00000368739.3
|
PGLYRP4
|
peptidoglycan recognition protein 4 |
| chr12_-_42877726 | 0.03 |
ENST00000548696.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr14_+_59100774 | 0.03 |
ENST00000556859.1
ENST00000421793.1 |
DACT1
|
dishevelled-binding antagonist of beta-catenin 1 |
| chr10_-_65028817 | 0.03 |
ENST00000542921.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr15_-_64385981 | 0.03 |
ENST00000557835.1
ENST00000380290.3 ENST00000559950.1 |
FAM96A
|
family with sequence similarity 96, member A |
| chr14_-_24551195 | 0.03 |
ENST00000560550.1
|
NRL
|
neural retina leucine zipper |
| chr15_+_67841330 | 0.03 |
ENST00000354498.5
|
MAP2K5
|
mitogen-activated protein kinase kinase 5 |
| chr22_-_29457832 | 0.03 |
ENST00000216071.4
|
C22orf31
|
chromosome 22 open reading frame 31 |
| chr2_+_182756615 | 0.03 |
ENST00000431877.2
ENST00000320370.7 |
SSFA2
|
sperm specific antigen 2 |
| chr12_-_71031185 | 0.03 |
ENST00000548122.1
ENST00000551525.1 ENST00000550358.1 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr3_+_189507523 | 0.03 |
ENST00000437221.1
ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63
|
tumor protein p63 |
| chr1_-_241683001 | 0.03 |
ENST00000366560.3
|
FH
|
fumarate hydratase |
| chrX_+_134887233 | 0.03 |
ENST00000443882.1
|
CT45A3
|
cancer/testis antigen family 45, member A3 |
| chr3_+_132379154 | 0.03 |
ENST00000468022.1
ENST00000473651.1 ENST00000494238.2 |
UBA5
|
ubiquitin-like modifier activating enzyme 5 |
| chr1_-_201915590 | 0.02 |
ENST00000367288.4
|
LMOD1
|
leiomodin 1 (smooth muscle) |
| chr2_-_197226875 | 0.02 |
ENST00000409111.1
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr2_-_109605663 | 0.02 |
ENST00000409271.1
ENST00000258443.2 ENST00000376651.1 |
EDAR
|
ectodysplasin A receptor |
| chr12_-_42877764 | 0.02 |
ENST00000455697.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr15_+_64386261 | 0.02 |
ENST00000560829.1
|
SNX1
|
sorting nexin 1 |
| chr2_+_68962014 | 0.02 |
ENST00000467265.1
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr1_-_94586651 | 0.02 |
ENST00000535735.1
ENST00000370225.3 |
ABCA4
|
ATP-binding cassette, sub-family A (ABC1), member 4 |
| chr15_+_91643442 | 0.02 |
ENST00000394232.1
|
SV2B
|
synaptic vesicle glycoprotein 2B |
| chr15_+_27111510 | 0.02 |
ENST00000335625.5
|
GABRA5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr10_+_124907638 | 0.02 |
ENST00000339992.3
|
HMX2
|
H6 family homeobox 2 |
| chr17_+_34640031 | 0.02 |
ENST00000339270.6
ENST00000482104.1 |
CCL4L2
|
chemokine (C-C motif) ligand 4-like 2 |
| chr6_+_31887761 | 0.01 |
ENST00000413154.1
|
C2
|
complement component 2 |
| chr14_+_56127960 | 0.01 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr1_+_78383813 | 0.01 |
ENST00000342754.5
|
NEXN
|
nexilin (F actin binding protein) |
| chr4_-_46391931 | 0.01 |
ENST00000381620.4
|
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2 |
| chr4_-_48116540 | 0.01 |
ENST00000506073.1
|
TXK
|
TXK tyrosine kinase |
| chr17_-_8770956 | 0.01 |
ENST00000311434.9
|
PIK3R6
|
phosphoinositide-3-kinase, regulatory subunit 6 |
| chr2_+_234526272 | 0.01 |
ENST00000373450.4
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr12_-_111358372 | 0.01 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr5_+_74011328 | 0.01 |
ENST00000513336.1
|
HEXB
|
hexosaminidase B (beta polypeptide) |
| chr8_+_107593198 | 0.01 |
ENST00000517686.1
|
OXR1
|
oxidation resistance 1 |
| chr2_+_103378472 | 0.01 |
ENST00000412401.2
|
TMEM182
|
transmembrane protein 182 |
| chrX_-_100546314 | 0.01 |
ENST00000356784.1
|
TAF7L
|
TAF7-like RNA polymerase II, TATA box binding protein (TBP)-associated factor, 50kDa |
| chr3_+_158787041 | 0.01 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr14_-_24733444 | 0.01 |
ENST00000560478.1
ENST00000560443.1 |
TGM1
|
transglutaminase 1 |
| chr7_-_76829125 | 0.01 |
ENST00000248598.5
|
FGL2
|
fibrinogen-like 2 |
| chr17_+_44588877 | 0.00 |
ENST00000576629.1
|
LRRC37A2
|
leucine rich repeat containing 37, member A2 |
| chr14_+_56127989 | 0.00 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr15_+_77308152 | 0.00 |
ENST00000559859.1
|
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1 |
| chrX_+_100878079 | 0.00 |
ENST00000471229.2
|
ARMCX3
|
armadillo repeat containing, X-linked 3 |
| chr2_+_197504278 | 0.00 |
ENST00000272831.7
ENST00000389175.4 ENST00000472405.2 ENST00000423093.2 |
CCDC150
|
coiled-coil domain containing 150 |
| chr7_+_55433131 | 0.00 |
ENST00000254770.2
|
LANCL2
|
LanC lantibiotic synthetase component C-like 2 (bacterial) |
Network of associatons between targets according to the STRING database.
First level regulatory network of STAT6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) isoquinoline alkaloid metabolic process(GO:0033076) |
| 0.1 | 0.4 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.1 | 0.3 | GO:1903565 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.1 | 0.3 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.1 | 0.4 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 0.3 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.1 | 0.3 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.1 | 0.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.2 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.2 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.0 | 0.4 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.1 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.2 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.3 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.2 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) regulation of stem cell division(GO:2000035) |
| 0.0 | 0.2 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.2 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.1 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.1 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.2 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.1 | GO:2000690 | regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.3 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.0 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 0.0 | 0.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.1 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.1 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.2 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 0.3 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.0 | 0.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.0 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.4 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |