Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
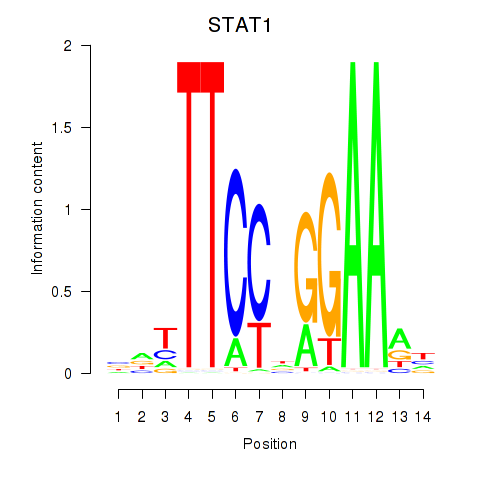
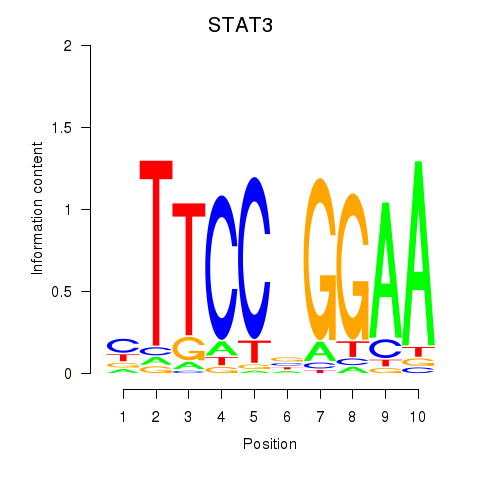
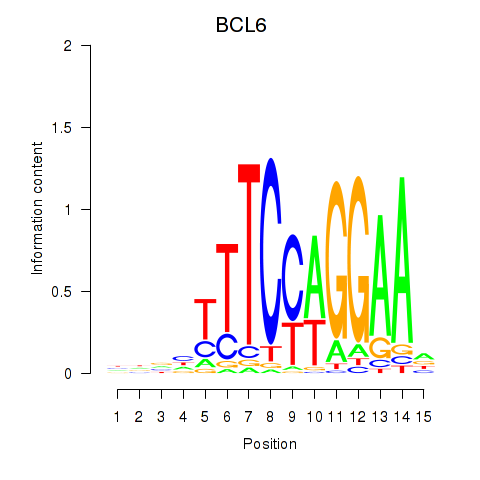
Results for STAT1_STAT3_BCL6
Z-value: 0.77
Transcription factors associated with STAT1_STAT3_BCL6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
STAT1
|
ENSG00000115415.14 | signal transducer and activator of transcription 1 |
|
STAT3
|
ENSG00000168610.10 | signal transducer and activator of transcription 3 |
|
BCL6
|
ENSG00000113916.13 | BCL6 transcription repressor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| STAT3 | hg19_v2_chr17_-_40540377_40540481 | -0.43 | 5.7e-01 | Click! |
| BCL6 | hg19_v2_chr3_-_187455680_187455732 | -0.30 | 7.0e-01 | Click! |
| STAT1 | hg19_v2_chr2_-_191878681_191878823 | -0.29 | 7.1e-01 | Click! |
Activity profile of STAT1_STAT3_BCL6 motif
Sorted Z-values of STAT1_STAT3_BCL6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_4717835 | 0.63 |
ENST00000599248.1
|
DPP9
|
dipeptidyl-peptidase 9 |
| chr2_+_87755054 | 0.44 |
ENST00000423846.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr19_-_51875894 | 0.41 |
ENST00000600427.1
ENST00000595217.1 ENST00000221978.5 |
NKG7
|
natural killer cell group 7 sequence |
| chr10_+_6625605 | 0.34 |
ENST00000414894.1
ENST00000449648.1 |
PRKCQ-AS1
|
PRKCQ antisense RNA 1 |
| chr17_+_2264983 | 0.30 |
ENST00000574650.1
|
SGSM2
|
small G protein signaling modulator 2 |
| chrX_-_154563889 | 0.28 |
ENST00000369449.2
ENST00000321926.4 |
CLIC2
|
chloride intracellular channel 2 |
| chr8_+_71485681 | 0.26 |
ENST00000391684.1
|
AC120194.1
|
AC120194.1 |
| chr14_+_76452090 | 0.24 |
ENST00000314067.6
ENST00000238628.6 ENST00000556742.1 |
IFT43
|
intraflagellar transport 43 homolog (Chlamydomonas) |
| chr17_-_46691990 | 0.22 |
ENST00000576562.1
|
HOXB8
|
homeobox B8 |
| chr19_+_35225121 | 0.21 |
ENST00000595708.1
ENST00000593781.1 |
ZNF181
|
zinc finger protein 181 |
| chr14_-_81408063 | 0.20 |
ENST00000557411.1
|
CEP128
|
centrosomal protein 128kDa |
| chr14_-_80678512 | 0.19 |
ENST00000553968.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chr19_+_47835404 | 0.19 |
ENST00000595464.1
|
C5AR2
|
complement component 5a receptor 2 |
| chr9_-_117880477 | 0.19 |
ENST00000534839.1
ENST00000340094.3 ENST00000535648.1 ENST00000346706.3 ENST00000345230.3 ENST00000350763.4 |
TNC
|
tenascin C |
| chr1_-_144994840 | 0.18 |
ENST00000369351.3
ENST00000369349.3 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr10_+_35484793 | 0.18 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr16_-_67260691 | 0.17 |
ENST00000447579.1
ENST00000393992.1 ENST00000424285.1 |
LRRC29
|
leucine rich repeat containing 29 |
| chr16_-_67517716 | 0.17 |
ENST00000290953.2
|
AGRP
|
agouti related protein homolog (mouse) |
| chr19_-_40324255 | 0.15 |
ENST00000593685.1
ENST00000600611.1 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr11_-_65363259 | 0.15 |
ENST00000342202.4
|
KCNK7
|
potassium channel, subfamily K, member 7 |
| chr17_+_6347729 | 0.15 |
ENST00000572447.1
|
FAM64A
|
family with sequence similarity 64, member A |
| chr16_-_67260901 | 0.14 |
ENST00000341546.3
ENST00000409509.1 ENST00000433915.1 ENST00000454102.2 |
LRRC29
AC040160.1
|
leucine rich repeat containing 29 Uncharacterized protein; cDNA FLJ57407, weakly similar to Mus musculus leucine rich repeat containing 29 (Lrrc29), mRNA |
| chr1_+_207262881 | 0.13 |
ENST00000451804.2
|
C4BPB
|
complement component 4 binding protein, beta |
| chr17_-_41132410 | 0.13 |
ENST00000409446.3
ENST00000453594.1 ENST00000409399.1 ENST00000421990.2 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr3_+_45430105 | 0.13 |
ENST00000430399.1
|
LARS2
|
leucyl-tRNA synthetase 2, mitochondrial |
| chr11_-_104817919 | 0.13 |
ENST00000533252.1
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr18_+_66382428 | 0.13 |
ENST00000578970.1
ENST00000582371.1 ENST00000584775.1 |
CCDC102B
|
coiled-coil domain containing 102B |
| chr19_-_40324767 | 0.12 |
ENST00000601972.1
ENST00000430012.2 ENST00000323039.5 ENST00000348817.3 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr6_-_83775442 | 0.12 |
ENST00000369745.5
|
UBE3D
|
ubiquitin protein ligase E3D |
| chr12_+_53443963 | 0.12 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr9_+_125796806 | 0.12 |
ENST00000373642.1
|
GPR21
|
G protein-coupled receptor 21 |
| chr17_+_76311791 | 0.12 |
ENST00000586321.1
|
AC061992.2
|
AC061992.2 |
| chr7_-_26578407 | 0.12 |
ENST00000242109.3
|
KIAA0087
|
KIAA0087 |
| chr17_-_42994283 | 0.12 |
ENST00000593179.1
|
GFAP
|
glial fibrillary acidic protein |
| chr17_-_46692287 | 0.12 |
ENST00000239144.4
|
HOXB8
|
homeobox B8 |
| chr12_+_56414851 | 0.12 |
ENST00000547167.1
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr8_-_102803163 | 0.12 |
ENST00000523645.1
ENST00000520346.1 ENST00000220931.6 ENST00000522448.1 ENST00000522951.1 ENST00000522252.1 ENST00000519098.1 |
NCALD
|
neurocalcin delta |
| chr2_-_204398141 | 0.12 |
ENST00000428637.1
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr21_+_17961006 | 0.11 |
ENST00000602323.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr3_-_112329110 | 0.11 |
ENST00000479368.1
|
CCDC80
|
coiled-coil domain containing 80 |
| chr10_-_128110441 | 0.11 |
ENST00000456514.1
|
LINC00601
|
long intergenic non-protein coding RNA 601 |
| chr5_+_667759 | 0.11 |
ENST00000594226.1
|
AC026740.1
|
Uncharacterized protein |
| chr10_-_90751038 | 0.11 |
ENST00000458159.1
ENST00000415557.1 ENST00000458208.1 |
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr1_-_230850043 | 0.10 |
ENST00000366667.4
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8) |
| chr19_-_58864848 | 0.10 |
ENST00000263100.3
|
A1BG
|
alpha-1-B glycoprotein |
| chr14_+_39944025 | 0.10 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr12_+_53443680 | 0.10 |
ENST00000314250.6
ENST00000451358.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr3_+_48264816 | 0.10 |
ENST00000296435.2
ENST00000576243.1 |
CAMP
|
cathelicidin antimicrobial peptide |
| chr7_-_87936195 | 0.09 |
ENST00000414498.1
ENST00000301959.5 ENST00000380079.4 |
STEAP4
|
STEAP family member 4 |
| chr8_-_27468842 | 0.09 |
ENST00000523500.1
|
CLU
|
clusterin |
| chr8_-_27468945 | 0.09 |
ENST00000405140.3
|
CLU
|
clusterin |
| chr15_+_84322827 | 0.09 |
ENST00000286744.5
ENST00000567476.1 |
ADAMTSL3
|
ADAMTS-like 3 |
| chr19_-_4831701 | 0.09 |
ENST00000248244.5
|
TICAM1
|
toll-like receptor adaptor molecule 1 |
| chr1_+_10534944 | 0.09 |
ENST00000356607.4
ENST00000538836.1 ENST00000491661.2 |
PEX14
|
peroxisomal biogenesis factor 14 |
| chr19_+_35773242 | 0.09 |
ENST00000222304.3
|
HAMP
|
hepcidin antimicrobial peptide |
| chr1_-_95846528 | 0.09 |
ENST00000427695.3
|
RP11-14O19.2
|
RP11-14O19.2 |
| chr6_+_32821924 | 0.09 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr6_+_139349903 | 0.09 |
ENST00000461027.1
|
ABRACL
|
ABRA C-terminal like |
| chr10_-_101841588 | 0.09 |
ENST00000370418.3
|
CPN1
|
carboxypeptidase N, polypeptide 1 |
| chr20_+_44637526 | 0.09 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr6_-_127840048 | 0.09 |
ENST00000467753.1
|
SOGA3
|
SOGA family member 3 |
| chr12_-_120687948 | 0.09 |
ENST00000458477.2
|
PXN
|
paxillin |
| chr12_+_56415100 | 0.09 |
ENST00000547791.1
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr17_-_28661065 | 0.09 |
ENST00000328886.4
ENST00000538566.2 |
TMIGD1
|
transmembrane and immunoglobulin domain containing 1 |
| chr4_-_140477353 | 0.09 |
ENST00000406354.1
ENST00000506866.2 |
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr15_+_86087267 | 0.09 |
ENST00000558166.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr11_+_46740730 | 0.09 |
ENST00000311907.5
ENST00000530231.1 ENST00000442468.1 |
F2
|
coagulation factor II (thrombin) |
| chr3_+_142842128 | 0.08 |
ENST00000483262.1
|
RP11-80H8.4
|
RP11-80H8.4 |
| chr22_-_50699701 | 0.08 |
ENST00000395780.1
|
MAPK12
|
mitogen-activated protein kinase 12 |
| chr11_-_36310958 | 0.08 |
ENST00000532705.1
ENST00000263401.5 ENST00000452374.2 |
COMMD9
|
COMM domain containing 9 |
| chr11_+_57531292 | 0.08 |
ENST00000524579.1
|
CTNND1
|
catenin (cadherin-associated protein), delta 1 |
| chr1_+_6845384 | 0.08 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr3_-_52864680 | 0.08 |
ENST00000406595.1
ENST00000485816.1 ENST00000434759.3 ENST00000346281.5 ENST00000266041.4 |
ITIH4
|
inter-alpha-trypsin inhibitor heavy chain family, member 4 |
| chr1_+_221051699 | 0.08 |
ENST00000366903.6
|
HLX
|
H2.0-like homeobox |
| chr19_+_17416609 | 0.08 |
ENST00000602206.1
|
MRPL34
|
mitochondrial ribosomal protein L34 |
| chr3_+_122399697 | 0.08 |
ENST00000494811.1
|
PARP14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr3_+_112930946 | 0.08 |
ENST00000462425.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr12_-_21928515 | 0.08 |
ENST00000537950.1
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr17_+_6347761 | 0.08 |
ENST00000250056.8
ENST00000571373.1 ENST00000570337.2 ENST00000572595.2 ENST00000576056.1 |
FAM64A
|
family with sequence similarity 64, member A |
| chr1_+_27248203 | 0.08 |
ENST00000321265.5
|
NUDC
|
nudC nuclear distribution protein |
| chr16_-_8955601 | 0.08 |
ENST00000569398.1
ENST00000568968.1 |
CARHSP1
|
calcium regulated heat stable protein 1, 24kDa |
| chr11_+_75479850 | 0.08 |
ENST00000376262.3
ENST00000604733.1 |
DGAT2
|
diacylglycerol O-acyltransferase 2 |
| chr19_+_17416457 | 0.08 |
ENST00000252602.1
|
MRPL34
|
mitochondrial ribosomal protein L34 |
| chr16_-_8955570 | 0.08 |
ENST00000567554.1
|
CARHSP1
|
calcium regulated heat stable protein 1, 24kDa |
| chr2_-_99797473 | 0.08 |
ENST00000409107.1
ENST00000289359.2 |
MITD1
|
MIT, microtubule interacting and transport, domain containing 1 |
| chr1_+_43613612 | 0.07 |
ENST00000335282.4
|
FAM183A
|
family with sequence similarity 183, member A |
| chr5_-_111093081 | 0.07 |
ENST00000453526.2
ENST00000509427.1 |
NREP
|
neuronal regeneration related protein |
| chr16_+_67261008 | 0.07 |
ENST00000304800.9
ENST00000563953.1 ENST00000565201.1 |
TMEM208
|
transmembrane protein 208 |
| chr1_-_31845914 | 0.07 |
ENST00000373713.2
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) |
| chrX_+_129473859 | 0.07 |
ENST00000424447.1
|
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr12_-_123187890 | 0.07 |
ENST00000328880.5
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chr9_-_117853297 | 0.07 |
ENST00000542877.1
ENST00000537320.1 ENST00000341037.4 |
TNC
|
tenascin C |
| chr9_-_89562104 | 0.07 |
ENST00000298743.7
|
GAS1
|
growth arrest-specific 1 |
| chr10_+_115511213 | 0.07 |
ENST00000361048.1
|
PLEKHS1
|
pleckstrin homology domain containing, family S member 1 |
| chr11_-_119999611 | 0.07 |
ENST00000529044.1
|
TRIM29
|
tripartite motif containing 29 |
| chr1_-_144994909 | 0.07 |
ENST00000369347.4
ENST00000369354.3 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr1_+_17906970 | 0.07 |
ENST00000375415.1
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr11_+_74303612 | 0.07 |
ENST00000527458.1
ENST00000532497.1 ENST00000530511.1 |
POLD3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr4_-_155511887 | 0.07 |
ENST00000302053.3
ENST00000403106.3 |
FGA
|
fibrinogen alpha chain |
| chr8_-_27469196 | 0.07 |
ENST00000546343.1
ENST00000560566.1 |
CLU
|
clusterin |
| chr12_+_56414795 | 0.07 |
ENST00000431367.2
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr2_+_87754887 | 0.07 |
ENST00000409054.1
ENST00000331944.6 ENST00000409139.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr6_-_45544507 | 0.07 |
ENST00000563807.1
|
RP1-166H4.2
|
RP1-166H4.2 |
| chr9_-_134151915 | 0.07 |
ENST00000372271.3
|
FAM78A
|
family with sequence similarity 78, member A |
| chr11_-_8290263 | 0.07 |
ENST00000428101.2
|
LMO1
|
LIM domain only 1 (rhombotin 1) |
| chr11_-_62609281 | 0.07 |
ENST00000525239.1
ENST00000538098.2 |
WDR74
|
WD repeat domain 74 |
| chr4_-_2758015 | 0.07 |
ENST00000510267.1
ENST00000503235.1 ENST00000315423.7 |
TNIP2
|
TNFAIP3 interacting protein 2 |
| chr14_+_22458631 | 0.07 |
ENST00000390444.1
|
TRAV16
|
T cell receptor alpha variable 16 |
| chr12_-_123201337 | 0.07 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr11_-_119999539 | 0.07 |
ENST00000541857.1
|
TRIM29
|
tripartite motif containing 29 |
| chr19_+_49467232 | 0.07 |
ENST00000599784.1
ENST00000594305.1 |
CTD-2639E6.9
|
CTD-2639E6.9 |
| chr4_+_68424434 | 0.07 |
ENST00000265404.2
ENST00000396225.1 |
STAP1
|
signal transducing adaptor family member 1 |
| chr19_+_42746927 | 0.07 |
ENST00000378108.1
|
AC006486.1
|
AC006486.1 |
| chr1_-_8000872 | 0.07 |
ENST00000377507.3
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr16_-_3030407 | 0.07 |
ENST00000431515.2
ENST00000574385.1 ENST00000576268.1 ENST00000574730.1 ENST00000575632.1 ENST00000573944.1 ENST00000262300.8 |
PKMYT1
|
protein kinase, membrane associated tyrosine/threonine 1 |
| chr2_-_84686552 | 0.07 |
ENST00000393868.2
|
SUCLG1
|
succinate-CoA ligase, alpha subunit |
| chr19_+_44331555 | 0.07 |
ENST00000590950.1
|
ZNF283
|
zinc finger protein 283 |
| chr2_-_99797390 | 0.07 |
ENST00000422537.2
|
MITD1
|
MIT, microtubule interacting and transport, domain containing 1 |
| chr12_+_27849378 | 0.07 |
ENST00000310791.2
|
REP15
|
RAB15 effector protein |
| chr1_+_59250815 | 0.07 |
ENST00000544621.1
ENST00000419531.2 |
RP4-794H19.2
|
long intergenic non-protein coding RNA 1135 |
| chr22_-_29457832 | 0.06 |
ENST00000216071.4
|
C22orf31
|
chromosome 22 open reading frame 31 |
| chr12_+_12224331 | 0.06 |
ENST00000396367.1
ENST00000266434.4 ENST00000396369.1 |
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr16_-_10652993 | 0.06 |
ENST00000536829.1
|
EMP2
|
epithelial membrane protein 2 |
| chr17_-_78428487 | 0.06 |
ENST00000562672.2
|
CTD-2526A2.2
|
CTD-2526A2.2 |
| chr11_+_123986069 | 0.06 |
ENST00000456829.2
ENST00000361352.5 ENST00000449321.1 ENST00000392748.1 ENST00000360334.4 ENST00000392744.4 |
VWA5A
|
von Willebrand factor A domain containing 5A |
| chr5_-_140013224 | 0.06 |
ENST00000498971.2
|
CD14
|
CD14 molecule |
| chr20_+_48909240 | 0.06 |
ENST00000371639.3
|
RP11-290F20.1
|
RP11-290F20.1 |
| chr12_-_49245936 | 0.06 |
ENST00000308025.3
|
DDX23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 |
| chr12_-_56120865 | 0.06 |
ENST00000548898.1
ENST00000552067.1 |
CD63
|
CD63 molecule |
| chr2_-_242089677 | 0.06 |
ENST00000405260.1
|
PASK
|
PAS domain containing serine/threonine kinase |
| chr1_-_226065330 | 0.06 |
ENST00000436966.1
|
TMEM63A
|
transmembrane protein 63A |
| chr7_+_2598628 | 0.06 |
ENST00000402050.2
ENST00000404984.1 ENST00000415271.2 ENST00000438376.2 |
IQCE
|
IQ motif containing E |
| chr17_-_19265982 | 0.06 |
ENST00000268841.6
ENST00000261499.4 ENST00000575478.1 |
B9D1
|
B9 protein domain 1 |
| chr5_-_119669160 | 0.06 |
ENST00000514240.1
|
CTC-552D5.1
|
CTC-552D5.1 |
| chr9_-_99180597 | 0.06 |
ENST00000375256.4
|
ZNF367
|
zinc finger protein 367 |
| chr22_-_31503490 | 0.06 |
ENST00000400299.2
|
SELM
|
Selenoprotein M |
| chr1_-_144995002 | 0.06 |
ENST00000369356.4
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr4_+_86396265 | 0.06 |
ENST00000395184.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr1_+_182584314 | 0.06 |
ENST00000566297.1
|
RP11-317P15.4
|
RP11-317P15.4 |
| chr11_+_5711010 | 0.06 |
ENST00000454828.1
|
TRIM22
|
tripartite motif containing 22 |
| chr11_+_73003824 | 0.06 |
ENST00000538328.1
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr12_-_56120838 | 0.06 |
ENST00000548160.1
|
CD63
|
CD63 molecule |
| chr16_+_48657361 | 0.06 |
ENST00000565072.1
|
RP11-42I10.1
|
RP11-42I10.1 |
| chr19_+_50145328 | 0.06 |
ENST00000360565.3
|
SCAF1
|
SR-related CTD-associated factor 1 |
| chr7_+_117120017 | 0.06 |
ENST00000003084.6
ENST00000454343.1 |
CFTR
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr14_-_76447494 | 0.06 |
ENST00000238682.3
|
TGFB3
|
transforming growth factor, beta 3 |
| chr6_+_27925019 | 0.06 |
ENST00000244623.1
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6 |
| chr17_-_38210644 | 0.06 |
ENST00000394128.2
ENST00000394127.2 ENST00000356271.3 ENST00000535071.2 ENST00000580885.1 ENST00000543759.2 ENST00000537674.2 ENST00000580517.1 ENST00000578161.1 |
MED24
|
mediator complex subunit 24 |
| chr19_-_12845467 | 0.06 |
ENST00000592273.1
ENST00000588213.1 |
C19orf43
|
chromosome 19 open reading frame 43 |
| chrX_+_77154935 | 0.06 |
ENST00000481445.1
|
COX7B
|
cytochrome c oxidase subunit VIIb |
| chr14_-_24615805 | 0.06 |
ENST00000560410.1
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr14_-_105647606 | 0.05 |
ENST00000392568.2
|
NUDT14
|
nudix (nucleoside diphosphate linked moiety X)-type motif 14 |
| chr19_-_12845550 | 0.05 |
ENST00000242784.4
|
C19orf43
|
chromosome 19 open reading frame 43 |
| chr17_-_76274572 | 0.05 |
ENST00000374945.1
|
RP11-219G17.4
|
RP11-219G17.4 |
| chr1_+_165864800 | 0.05 |
ENST00000469256.2
|
UCK2
|
uridine-cytidine kinase 2 |
| chr2_-_166651191 | 0.05 |
ENST00000392701.3
|
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) |
| chr17_-_27038063 | 0.05 |
ENST00000439862.3
|
PROCA1
|
protein interacting with cyclin A1 |
| chr19_-_45927622 | 0.05 |
ENST00000300853.3
ENST00000589165.1 |
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr11_+_7506837 | 0.05 |
ENST00000528758.1
|
OLFML1
|
olfactomedin-like 1 |
| chr15_-_72462384 | 0.05 |
ENST00000568594.1
|
GRAMD2
|
GRAM domain containing 2 |
| chr2_-_197675000 | 0.05 |
ENST00000342506.2
|
C2orf66
|
chromosome 2 open reading frame 66 |
| chrX_+_100646190 | 0.05 |
ENST00000471855.1
|
RPL36A
|
ribosomal protein L36a |
| chr10_-_6622201 | 0.05 |
ENST00000539722.1
ENST00000397176.2 |
PRKCQ
|
protein kinase C, theta |
| chr16_+_70332956 | 0.05 |
ENST00000288071.6
ENST00000393657.2 ENST00000355992.3 ENST00000567706.1 |
DDX19B
RP11-529K1.3
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 19B Uncharacterized protein |
| chr19_-_55672037 | 0.05 |
ENST00000588076.1
|
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr17_+_76356516 | 0.05 |
ENST00000592569.1
|
RP11-806H10.4
|
RP11-806H10.4 |
| chr7_+_35756092 | 0.05 |
ENST00000458087.3
|
AC018647.3
|
AC018647.3 |
| chr2_+_87754989 | 0.05 |
ENST00000409898.2
ENST00000419680.2 ENST00000414584.1 ENST00000455131.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr20_+_42965626 | 0.05 |
ENST00000217043.2
|
R3HDML
|
R3H domain containing-like |
| chr4_-_185820602 | 0.05 |
ENST00000515864.1
ENST00000507183.1 |
RP11-701P16.4
|
long intergenic non-protein coding RNA 1093 |
| chr9_+_139702363 | 0.05 |
ENST00000371663.4
ENST00000371671.4 ENST00000311502.7 |
RABL6
|
RAB, member RAS oncogene family-like 6 |
| chr19_-_49339732 | 0.05 |
ENST00000599157.1
|
HSD17B14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr19_-_44809121 | 0.05 |
ENST00000591609.1
ENST00000589799.1 ENST00000291182.4 ENST00000589248.1 |
ZNF235
|
zinc finger protein 235 |
| chr6_+_108487245 | 0.05 |
ENST00000368986.4
|
NR2E1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr5_-_64777685 | 0.05 |
ENST00000536360.1
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
| chr5_-_64777733 | 0.05 |
ENST00000381055.3
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
| chr2_-_166651152 | 0.05 |
ENST00000431484.1
ENST00000412248.1 |
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) |
| chr12_-_15082050 | 0.05 |
ENST00000540097.1
|
ERP27
|
endoplasmic reticulum protein 27 |
| chr11_-_111649015 | 0.05 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chrX_-_48931648 | 0.05 |
ENST00000376386.3
ENST00000376390.4 |
PRAF2
|
PRA1 domain family, member 2 |
| chr1_+_244227632 | 0.05 |
ENST00000598000.1
|
AL590483.1
|
Uncharacterized protein; cDNA FLJ42623 fis, clone BRACE3015894 |
| chr1_+_151138500 | 0.05 |
ENST00000368905.4
|
SCNM1
|
sodium channel modifier 1 |
| chr6_-_26235206 | 0.05 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr4_-_140222358 | 0.05 |
ENST00000505036.1
ENST00000544855.1 ENST00000539002.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr11_-_111782696 | 0.05 |
ENST00000227251.3
ENST00000526180.1 |
CRYAB
|
crystallin, alpha B |
| chr7_-_83278322 | 0.05 |
ENST00000307792.3
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr1_+_155278625 | 0.05 |
ENST00000368356.4
ENST00000356657.6 |
FDPS
|
farnesyl diphosphate synthase |
| chr14_-_38036271 | 0.05 |
ENST00000556024.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr20_-_62582475 | 0.05 |
ENST00000369908.5
|
UCKL1
|
uridine-cytidine kinase 1-like 1 |
| chr8_-_144886321 | 0.05 |
ENST00000526832.1
|
SCRIB
|
scribbled planar cell polarity protein |
| chr1_-_144995074 | 0.05 |
ENST00000534536.1
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr7_-_44613494 | 0.05 |
ENST00000431640.1
ENST00000258772.5 |
DDX56
|
DEAD (Asp-Glu-Ala-Asp) box helicase 56 |
| chr20_-_43280361 | 0.04 |
ENST00000372874.4
|
ADA
|
adenosine deaminase |
| chr7_-_100881041 | 0.04 |
ENST00000412417.1
ENST00000414035.1 |
CLDN15
|
claudin 15 |
| chr12_+_56618102 | 0.04 |
ENST00000267023.4
ENST00000380198.2 ENST00000341463.5 |
NABP2
|
nucleic acid binding protein 2 |
| chr14_+_24779376 | 0.04 |
ENST00000530080.1
|
LTB4R2
|
leukotriene B4 receptor 2 |
| chr5_-_111093340 | 0.04 |
ENST00000508870.1
|
NREP
|
neuronal regeneration related protein |
| chr7_-_149158187 | 0.04 |
ENST00000247930.4
|
ZNF777
|
zinc finger protein 777 |
| chr6_+_43484760 | 0.04 |
ENST00000372389.3
ENST00000372344.2 ENST00000304004.3 ENST00000423780.1 |
POLR1C
|
polymerase (RNA) I polypeptide C, 30kDa |
| chr19_+_15783879 | 0.04 |
ENST00000551607.1
|
CYP4F12
|
cytochrome P450, family 4, subfamily F, polypeptide 12 |
Network of associatons between targets according to the STRING database.
First level regulatory network of STAT1_STAT3_BCL6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.1 | 0.3 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.1 | 0.2 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 | 0.2 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.0 | 0.1 | GO:1903410 | lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine import into cell(GO:1903410) |
| 0.0 | 0.1 | GO:0042704 | detection of oxygen(GO:0003032) uterine wall breakdown(GO:0042704) |
| 0.0 | 0.1 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 | 0.1 | GO:1990641 | cellular response to bile acid(GO:1903413) response to iron ion starvation(GO:1990641) |
| 0.0 | 0.0 | GO:0014839 | myoblast migration involved in skeletal muscle regeneration(GO:0014839) |
| 0.0 | 0.1 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.2 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.1 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.1 | GO:1903980 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) positive regulation of microglial cell activation(GO:1903980) |
| 0.0 | 0.1 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.2 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.1 | GO:0033383 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.0 | 0.3 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.0 | 0.0 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.0 | 0.0 | GO:0002605 | negative regulation of dendritic cell antigen processing and presentation(GO:0002605) regulation of cytotoxic T cell degranulation(GO:0043317) negative regulation of cytotoxic T cell degranulation(GO:0043318) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.0 | 0.2 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.1 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.0 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.1 | GO:2001183 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.1 | GO:0052214 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.0 | 0.1 | GO:0035712 | T-helper 2 cell activation(GO:0035712) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.0 | 0.1 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.0 | 0.0 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.0 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.0 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.1 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.1 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.1 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.0 | 0.0 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.0 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.0 | 0.1 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.2 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.0 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.0 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.0 | 0.0 | GO:0051796 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.0 | 0.0 | GO:1902824 | positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.0 | 0.0 | GO:0035668 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.0 | 0.1 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.1 | GO:0045359 | positive regulation of interferon-beta biosynthetic process(GO:0045359) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.2 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.2 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.0 | GO:0000806 | Y chromosome(GO:0000806) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.1 | 0.2 | GO:0004823 | leucine-tRNA ligase activity(GO:0004823) |
| 0.0 | 0.1 | GO:0004952 | dopamine neurotransmitter receptor activity(GO:0004952) |
| 0.0 | 0.2 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.1 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.1 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.0 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.0 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) leukotriene receptor activity(GO:0004974) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |