Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
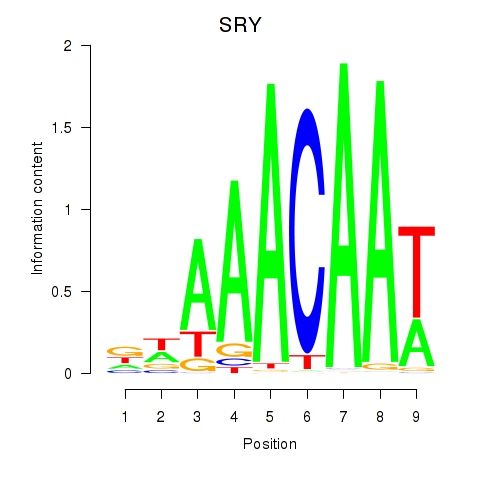
Results for SRY
Z-value: 0.99
Transcription factors associated with SRY
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SRY
|
ENSG00000184895.6 | sex determining region Y |
Activity profile of SRY motif
Sorted Z-values of SRY motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_115312766 | 0.91 |
ENST00000351270.3
|
HABP2
|
hyaluronan binding protein 2 |
| chr17_-_57229155 | 0.82 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr3_+_178866199 | 0.78 |
ENST00000263967.3
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr1_-_93645818 | 0.69 |
ENST00000370280.1
ENST00000479918.1 |
TMED5
|
transmembrane emp24 protein transport domain containing 5 |
| chr4_+_165675197 | 0.65 |
ENST00000515485.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr12_+_95611536 | 0.63 |
ENST00000549002.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr13_+_110958124 | 0.61 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr2_+_162101247 | 0.61 |
ENST00000439050.1
ENST00000436506.1 |
AC009299.3
|
AC009299.3 |
| chr20_-_3762087 | 0.57 |
ENST00000379756.3
|
SPEF1
|
sperm flagellar 1 |
| chr10_-_99094458 | 0.54 |
ENST00000371019.2
|
FRAT2
|
frequently rearranged in advanced T-cell lymphomas 2 |
| chr3_-_123339343 | 0.54 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr4_-_76598296 | 0.54 |
ENST00000395719.3
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr1_+_97188188 | 0.51 |
ENST00000541987.1
|
PTBP2
|
polypyrimidine tract binding protein 2 |
| chr12_+_50479101 | 0.50 |
ENST00000551966.1
|
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr10_+_115312825 | 0.50 |
ENST00000537906.1
ENST00000541666.1 |
HABP2
|
hyaluronan binding protein 2 |
| chr5_-_111091948 | 0.49 |
ENST00000447165.2
|
NREP
|
neuronal regeneration related protein |
| chr17_-_49124230 | 0.48 |
ENST00000510283.1
ENST00000510855.1 |
SPAG9
|
sperm associated antigen 9 |
| chr1_+_205682497 | 0.47 |
ENST00000598338.1
|
AC119673.1
|
AC119673.1 |
| chr14_-_92414055 | 0.46 |
ENST00000342058.4
|
FBLN5
|
fibulin 5 |
| chr4_+_76871883 | 0.45 |
ENST00000599764.1
|
AC110615.1
|
Uncharacterized protein |
| chr2_-_55237484 | 0.45 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chr14_-_92413353 | 0.45 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr2_-_157198860 | 0.45 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr1_-_204436344 | 0.44 |
ENST00000367184.2
|
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr14_+_38065052 | 0.44 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr3_+_142342228 | 0.44 |
ENST00000337777.3
|
PLS1
|
plastin 1 |
| chr10_-_14613968 | 0.44 |
ENST00000488576.1
ENST00000472095.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr18_+_72922710 | 0.43 |
ENST00000322038.5
|
TSHZ1
|
teashirt zinc finger homeobox 1 |
| chr17_-_46671323 | 0.43 |
ENST00000239151.5
|
HOXB5
|
homeobox B5 |
| chr17_+_76311791 | 0.43 |
ENST00000586321.1
|
AC061992.2
|
AC061992.2 |
| chr16_+_53920795 | 0.43 |
ENST00000431610.2
ENST00000460382.1 |
FTO
|
fat mass and obesity associated |
| chr20_-_52612705 | 0.43 |
ENST00000434986.2
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chrX_-_106959631 | 0.42 |
ENST00000486554.1
ENST00000372390.4 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr4_+_24661479 | 0.41 |
ENST00000569621.1
|
RP11-496D24.2
|
RP11-496D24.2 |
| chr15_-_54025300 | 0.41 |
ENST00000559418.1
|
WDR72
|
WD repeat domain 72 |
| chr14_-_92413727 | 0.40 |
ENST00000267620.10
|
FBLN5
|
fibulin 5 |
| chr3_-_28389922 | 0.39 |
ENST00000415852.1
|
AZI2
|
5-azacytidine induced 2 |
| chr1_-_245026388 | 0.39 |
ENST00000440865.1
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) |
| chr4_-_76598326 | 0.37 |
ENST00000503660.1
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr3_-_28390581 | 0.37 |
ENST00000479665.1
|
AZI2
|
5-azacytidine induced 2 |
| chr3_-_145940214 | 0.36 |
ENST00000481701.1
|
PLSCR4
|
phospholipid scramblase 4 |
| chr10_-_14614311 | 0.36 |
ENST00000479731.1
ENST00000468492.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr15_+_98503922 | 0.36 |
ENST00000268042.6
|
ARRDC4
|
arrestin domain containing 4 |
| chr9_-_123476612 | 0.35 |
ENST00000426959.1
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr12_-_102591604 | 0.35 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr1_-_53018654 | 0.35 |
ENST00000257177.4
ENST00000355809.4 ENST00000528642.1 ENST00000470626.1 ENST00000371544.3 |
ZCCHC11
|
zinc finger, CCHC domain containing 11 |
| chr12_-_121973974 | 0.35 |
ENST00000538379.1
ENST00000541318.1 ENST00000541511.1 |
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr1_-_108231101 | 0.35 |
ENST00000544443.1
ENST00000415432.2 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr7_+_30185406 | 0.34 |
ENST00000324489.5
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr3_-_57233966 | 0.33 |
ENST00000473921.1
ENST00000295934.3 |
HESX1
|
HESX homeobox 1 |
| chr20_-_52612468 | 0.33 |
ENST00000422805.1
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr19_-_47734448 | 0.32 |
ENST00000439096.2
|
BBC3
|
BCL2 binding component 3 |
| chr4_-_103746924 | 0.31 |
ENST00000505207.1
ENST00000502404.1 ENST00000507845.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr1_-_39395165 | 0.31 |
ENST00000372985.3
|
RHBDL2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr5_+_137673200 | 0.30 |
ENST00000434981.2
|
FAM53C
|
family with sequence similarity 53, member C |
| chr9_-_123476719 | 0.30 |
ENST00000373930.3
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr3_+_112930306 | 0.30 |
ENST00000495514.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr4_-_103747011 | 0.30 |
ENST00000350435.7
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr2_-_160473114 | 0.30 |
ENST00000392783.2
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr10_+_111985713 | 0.30 |
ENST00000239007.7
|
MXI1
|
MAX interactor 1, dimerization protein |
| chrM_+_8366 | 0.29 |
ENST00000361851.1
|
MT-ATP8
|
mitochondrially encoded ATP synthase 8 |
| chr6_-_111888474 | 0.29 |
ENST00000368735.1
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr16_-_15736881 | 0.29 |
ENST00000540441.2
|
KIAA0430
|
KIAA0430 |
| chr18_+_3466248 | 0.29 |
ENST00000581029.1
ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4
|
RP11-838N2.4 |
| chr14_-_58893832 | 0.29 |
ENST00000556007.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr3_+_28390637 | 0.29 |
ENST00000420223.1
ENST00000383768.2 |
ZCWPW2
|
zinc finger, CW type with PWWP domain 2 |
| chr11_-_125351481 | 0.28 |
ENST00000577924.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr9_-_14314518 | 0.28 |
ENST00000397581.2
|
NFIB
|
nuclear factor I/B |
| chr1_+_84630053 | 0.26 |
ENST00000394838.2
ENST00000370682.3 ENST00000432111.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chrX_+_84499038 | 0.26 |
ENST00000373165.3
|
ZNF711
|
zinc finger protein 711 |
| chrX_+_123095546 | 0.26 |
ENST00000371157.3
ENST00000371145.3 ENST00000371144.3 |
STAG2
|
stromal antigen 2 |
| chr9_+_116343192 | 0.26 |
ENST00000471324.2
|
RGS3
|
regulator of G-protein signaling 3 |
| chr1_-_40367668 | 0.26 |
ENST00000397332.2
ENST00000429311.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr10_+_111967345 | 0.26 |
ENST00000332674.5
ENST00000453116.1 |
MXI1
|
MAX interactor 1, dimerization protein |
| chr4_-_140477928 | 0.25 |
ENST00000274031.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr18_+_46065570 | 0.25 |
ENST00000591412.1
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr3_-_11610255 | 0.24 |
ENST00000424529.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr6_-_79787902 | 0.24 |
ENST00000275034.4
|
PHIP
|
pleckstrin homology domain interacting protein |
| chr14_-_23288930 | 0.24 |
ENST00000554517.1
ENST00000285850.7 ENST00000397529.2 ENST00000555702.1 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr3_-_123339418 | 0.23 |
ENST00000583087.1
|
MYLK
|
myosin light chain kinase |
| chr11_+_28724129 | 0.23 |
ENST00000513853.1
|
RP11-115J23.1
|
RP11-115J23.1 |
| chr17_+_8924837 | 0.23 |
ENST00000173229.2
|
NTN1
|
netrin 1 |
| chr7_+_134551583 | 0.23 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr4_-_103746683 | 0.23 |
ENST00000504211.1
ENST00000508476.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr3_+_189507432 | 0.23 |
ENST00000354600.5
|
TP63
|
tumor protein p63 |
| chr4_-_83280767 | 0.23 |
ENST00000514671.1
|
HNRNPD
|
heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) |
| chr9_-_14314566 | 0.23 |
ENST00000397579.2
|
NFIB
|
nuclear factor I/B |
| chr14_-_58894223 | 0.22 |
ENST00000555593.1
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr1_+_93645314 | 0.22 |
ENST00000343253.7
|
CCDC18
|
coiled-coil domain containing 18 |
| chr2_+_27070964 | 0.22 |
ENST00000288699.6
|
DPYSL5
|
dihydropyrimidinase-like 5 |
| chr18_+_20715416 | 0.22 |
ENST00000580153.1
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1 |
| chr10_-_14614095 | 0.21 |
ENST00000482277.1
ENST00000378462.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr4_-_111120334 | 0.21 |
ENST00000503885.1
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr17_-_46716647 | 0.21 |
ENST00000608940.1
|
RP11-357H14.17
|
RP11-357H14.17 |
| chr6_+_108882069 | 0.21 |
ENST00000406360.1
|
FOXO3
|
forkhead box O3 |
| chr1_+_100316041 | 0.21 |
ENST00000370165.3
ENST00000370163.3 ENST00000294724.4 |
AGL
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
| chr1_+_33116765 | 0.20 |
ENST00000544435.1
ENST00000373485.1 ENST00000458695.2 ENST00000490500.1 ENST00000445722.2 |
RBBP4
|
retinoblastoma binding protein 4 |
| chr12_-_56352368 | 0.20 |
ENST00000549404.1
|
PMEL
|
premelanosome protein |
| chr7_+_7811992 | 0.20 |
ENST00000406829.1
|
RPA3-AS1
|
RPA3 antisense RNA 1 |
| chr10_+_111985837 | 0.20 |
ENST00000393134.1
|
MXI1
|
MAX interactor 1, dimerization protein |
| chr17_-_70053866 | 0.19 |
ENST00000540802.1
|
RP11-84E24.2
|
RP11-84E24.2 |
| chr14_-_58894332 | 0.19 |
ENST00000395159.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr4_+_124571409 | 0.19 |
ENST00000514823.1
ENST00000511919.1 ENST00000508111.1 |
RP11-93L9.1
|
long intergenic non-protein coding RNA 1091 |
| chr14_-_58893876 | 0.19 |
ENST00000555097.1
ENST00000555404.1 |
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr4_-_105416039 | 0.18 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr6_+_122720681 | 0.18 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chr17_+_76037081 | 0.18 |
ENST00000588549.1
|
TNRC6C
|
trinucleotide repeat containing 6C |
| chr12_+_50478977 | 0.18 |
ENST00000381513.4
|
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr4_-_76598544 | 0.18 |
ENST00000515457.1
ENST00000357854.3 |
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr9_-_39239171 | 0.18 |
ENST00000358144.2
|
CNTNAP3
|
contactin associated protein-like 3 |
| chr8_+_57124245 | 0.18 |
ENST00000521831.1
ENST00000355315.3 ENST00000303759.3 ENST00000517636.1 ENST00000517933.1 ENST00000518801.1 ENST00000523975.1 ENST00000396723.5 ENST00000523061.1 ENST00000521524.1 |
CHCHD7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr15_-_70994612 | 0.18 |
ENST00000558758.1
ENST00000379983.2 ENST00000560441.1 |
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr1_-_219615984 | 0.18 |
ENST00000420762.1
|
RP11-95P13.1
|
RP11-95P13.1 |
| chr10_-_14614122 | 0.17 |
ENST00000378465.3
ENST00000452706.2 ENST00000378458.2 |
FAM107B
|
family with sequence similarity 107, member B |
| chrX_+_84499081 | 0.17 |
ENST00000276123.3
|
ZNF711
|
zinc finger protein 711 |
| chr13_+_113633620 | 0.17 |
ENST00000421756.1
ENST00000375601.3 |
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr18_+_46065483 | 0.17 |
ENST00000382998.4
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr2_+_86668464 | 0.17 |
ENST00000409064.1
|
KDM3A
|
lysine (K)-specific demethylase 3A |
| chr1_-_160231451 | 0.17 |
ENST00000495887.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr12_-_2966193 | 0.17 |
ENST00000382678.3
|
AC005841.1
|
Uncharacterized protein ENSP00000372125 |
| chr5_-_131132614 | 0.17 |
ENST00000307968.7
ENST00000307954.8 |
FNIP1
|
folliculin interacting protein 1 |
| chrX_+_123095890 | 0.17 |
ENST00000435215.1
|
STAG2
|
stromal antigen 2 |
| chr3_-_135913298 | 0.16 |
ENST00000434835.2
|
MSL2
|
male-specific lethal 2 homolog (Drosophila) |
| chr15_-_55562451 | 0.16 |
ENST00000568803.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chrX_+_123095860 | 0.15 |
ENST00000428941.1
|
STAG2
|
stromal antigen 2 |
| chr5_+_110073853 | 0.15 |
ENST00000513807.1
ENST00000509442.2 |
SLC25A46
|
solute carrier family 25, member 46 |
| chr14_-_89960395 | 0.15 |
ENST00000555034.1
ENST00000553904.1 |
FOXN3
|
forkhead box N3 |
| chr3_-_28390298 | 0.15 |
ENST00000457172.1
|
AZI2
|
5-azacytidine induced 2 |
| chr15_+_59665194 | 0.15 |
ENST00000560394.1
|
FAM81A
|
family with sequence similarity 81, member A |
| chr15_+_90319557 | 0.15 |
ENST00000341735.3
|
MESP2
|
mesoderm posterior 2 homolog (mouse) |
| chr12_-_12674032 | 0.15 |
ENST00000298573.4
|
DUSP16
|
dual specificity phosphatase 16 |
| chr21_-_19191703 | 0.14 |
ENST00000284881.4
ENST00000400559.3 ENST00000400558.3 |
C21orf91
|
chromosome 21 open reading frame 91 |
| chr1_+_235490659 | 0.14 |
ENST00000488594.1
|
GGPS1
|
geranylgeranyl diphosphate synthase 1 |
| chr7_+_142985308 | 0.14 |
ENST00000310447.5
|
CASP2
|
caspase 2, apoptosis-related cysteine peptidase |
| chr12_-_53994805 | 0.14 |
ENST00000328463.7
|
ATF7
|
activating transcription factor 7 |
| chr16_+_2588012 | 0.14 |
ENST00000354836.5
ENST00000389224.3 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr4_+_165675269 | 0.14 |
ENST00000507311.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr15_+_59397275 | 0.14 |
ENST00000288207.2
|
CCNB2
|
cyclin B2 |
| chr11_-_10879572 | 0.14 |
ENST00000413761.2
|
ZBED5
|
zinc finger, BED-type containing 5 |
| chr2_+_162087577 | 0.14 |
ENST00000439442.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr11_+_120255997 | 0.14 |
ENST00000532993.1
|
ARHGEF12
|
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr4_-_103266626 | 0.13 |
ENST00000356736.4
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr16_+_2587998 | 0.13 |
ENST00000441549.3
ENST00000268673.7 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr3_-_141868293 | 0.13 |
ENST00000317104.7
ENST00000494358.1 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr14_+_67831576 | 0.13 |
ENST00000555876.1
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
| chr12_-_122985067 | 0.13 |
ENST00000540586.1
ENST00000543897.1 |
ZCCHC8
|
zinc finger, CCHC domain containing 8 |
| chr5_+_138677515 | 0.13 |
ENST00000265192.4
ENST00000511706.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr11_-_10879593 | 0.13 |
ENST00000528289.1
ENST00000432999.2 |
ZBED5
|
zinc finger, BED-type containing 5 |
| chr12_+_50479109 | 0.13 |
ENST00000550477.1
|
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr17_-_46703826 | 0.13 |
ENST00000550387.1
ENST00000311177.5 |
HOXB9
|
homeobox B9 |
| chr8_+_85095553 | 0.12 |
ENST00000521268.1
|
RALYL
|
RALY RNA binding protein-like |
| chr16_+_53469525 | 0.12 |
ENST00000544405.2
|
RBL2
|
retinoblastoma-like 2 (p130) |
| chr11_+_57529234 | 0.12 |
ENST00000360682.6
ENST00000361796.4 ENST00000529526.1 ENST00000426142.2 ENST00000399050.4 ENST00000361391.6 ENST00000361332.4 ENST00000532463.1 ENST00000529986.1 ENST00000358694.6 ENST00000532787.1 ENST00000533667.1 ENST00000532649.1 ENST00000528621.1 ENST00000530748.1 ENST00000428599.2 ENST00000527467.1 ENST00000528232.1 ENST00000531014.1 ENST00000526772.1 ENST00000529873.1 ENST00000525902.1 ENST00000532844.1 ENST00000526357.1 ENST00000530094.1 ENST00000415361.2 ENST00000532245.1 ENST00000534579.1 ENST00000526938.1 |
CTNND1
|
catenin (cadherin-associated protein), delta 1 |
| chr4_-_140477910 | 0.12 |
ENST00000404104.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr2_+_191273052 | 0.12 |
ENST00000417958.1
ENST00000432036.1 ENST00000392328.1 |
MFSD6
|
major facilitator superfamily domain containing 6 |
| chr4_-_164534657 | 0.12 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr2_+_173955327 | 0.12 |
ENST00000422149.1
|
MLTK
|
Mitogen-activated protein kinase kinase kinase MLT |
| chrX_+_78003204 | 0.12 |
ENST00000435339.3
ENST00000514744.1 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chr4_-_111120132 | 0.12 |
ENST00000506625.1
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr16_+_2587965 | 0.12 |
ENST00000342085.4
ENST00000566659.1 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr14_+_53019993 | 0.12 |
ENST00000542169.2
ENST00000555622.1 |
GPR137C
|
G protein-coupled receptor 137C |
| chr6_-_152489484 | 0.11 |
ENST00000354674.4
ENST00000539504.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr9_+_84304628 | 0.11 |
ENST00000437181.1
|
RP11-154D17.1
|
RP11-154D17.1 |
| chr2_-_37899323 | 0.11 |
ENST00000295324.3
ENST00000457889.1 |
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr7_+_29519486 | 0.11 |
ENST00000409041.4
|
CHN2
|
chimerin 2 |
| chr5_-_131132658 | 0.11 |
ENST00000514667.1
ENST00000511848.1 ENST00000510461.1 |
CTC-432M15.3
FNIP1
|
Folliculin-interacting protein 1 folliculin interacting protein 1 |
| chrX_-_16887963 | 0.11 |
ENST00000380084.4
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr10_+_99079008 | 0.11 |
ENST00000371021.3
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas |
| chr15_+_90544532 | 0.11 |
ENST00000268154.4
|
ZNF710
|
zinc finger protein 710 |
| chr2_+_109204743 | 0.11 |
ENST00000332345.6
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr6_+_31082603 | 0.11 |
ENST00000259881.9
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chr1_+_200011711 | 0.11 |
ENST00000544748.1
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr14_-_53019211 | 0.11 |
ENST00000557374.1
ENST00000281741.4 |
TXNDC16
|
thioredoxin domain containing 16 |
| chr1_-_40367530 | 0.11 |
ENST00000372816.2
ENST00000372815.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr3_+_189507460 | 0.10 |
ENST00000434928.1
|
TP63
|
tumor protein p63 |
| chr15_-_70390213 | 0.10 |
ENST00000557997.1
ENST00000317509.8 ENST00000442299.2 |
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr2_+_181845843 | 0.10 |
ENST00000602710.1
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr2_-_118943930 | 0.10 |
ENST00000449075.1
ENST00000414886.1 ENST00000449819.1 |
AC093901.1
|
AC093901.1 |
| chr18_+_11851383 | 0.10 |
ENST00000526991.2
|
CHMP1B
|
charged multivesicular body protein 1B |
| chr5_+_138678131 | 0.09 |
ENST00000394795.2
ENST00000510080.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chrX_+_9880412 | 0.09 |
ENST00000418909.2
|
SHROOM2
|
shroom family member 2 |
| chr3_-_28390120 | 0.09 |
ENST00000334100.6
|
AZI2
|
5-azacytidine induced 2 |
| chr21_+_17792672 | 0.09 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr8_+_21777159 | 0.09 |
ENST00000434536.1
ENST00000252512.9 |
XPO7
|
exportin 7 |
| chr12_-_92539614 | 0.09 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr20_-_22566089 | 0.09 |
ENST00000377115.4
|
FOXA2
|
forkhead box A2 |
| chr15_-_52263937 | 0.09 |
ENST00000315141.5
ENST00000299601.5 |
LEO1
|
Leo1, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) |
| chr2_+_181845763 | 0.09 |
ENST00000602499.1
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr1_+_61547405 | 0.08 |
ENST00000371189.4
|
NFIA
|
nuclear factor I/A |
| chr4_+_41614909 | 0.08 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr4_+_170581213 | 0.08 |
ENST00000507875.1
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr12_-_122985494 | 0.08 |
ENST00000336229.4
|
ZCCHC8
|
zinc finger, CCHC domain containing 8 |
| chr2_+_120517174 | 0.08 |
ENST00000263708.2
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr18_+_55712915 | 0.08 |
ENST00000592846.1
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr14_-_55878538 | 0.08 |
ENST00000247178.5
|
ATG14
|
autophagy related 14 |
| chr13_-_114843416 | 0.08 |
ENST00000389544.4
|
RASA3
|
RAS p21 protein activator 3 |
| chr20_-_35492048 | 0.08 |
ENST00000237536.4
|
SOGA1
|
suppressor of glucose, autophagy associated 1 |
| chr17_-_73511584 | 0.07 |
ENST00000321617.3
|
CASKIN2
|
CASK interacting protein 2 |
| chr16_-_4323015 | 0.07 |
ENST00000204517.6
|
TFAP4
|
transcription factor AP-4 (activating enhancer binding protein 4) |
| chrM_-_14670 | 0.07 |
ENST00000361681.2
|
MT-ND6
|
mitochondrially encoded NADH dehydrogenase 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SRY
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 | 0.4 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.1 | 0.4 | GO:0042245 | RNA repair(GO:0042245) |
| 0.1 | 1.1 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 0.8 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 1.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.9 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.4 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 0.3 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.2 | GO:1904586 | response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) hepatocyte dedifferentiation(GO:1990828) |
| 0.1 | 0.2 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 0.3 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.1 | 0.5 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 1.1 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 | 0.3 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 0.5 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.1 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.4 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.3 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.6 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.5 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.8 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.2 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.1 | GO:0045208 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.0 | 0.4 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.4 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.2 | GO:0007290 | spermatid nucleus elongation(GO:0007290) formaldehyde metabolic process(GO:0046292) |
| 0.0 | 0.7 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.7 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.3 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.4 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.2 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.4 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.6 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.2 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.2 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 | 0.7 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.0 | 0.2 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.3 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.2 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 1.0 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.4 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.0 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.0 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.1 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.0 | GO:0048687 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 1.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.8 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.6 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.8 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) astrocyte end-foot(GO:0097450) |
| 0.0 | 0.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.4 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.5 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.1 | 0.4 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.1 | 0.8 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.4 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.4 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 1.0 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 1.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.0 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.4 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.8 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.8 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.6 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |