Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
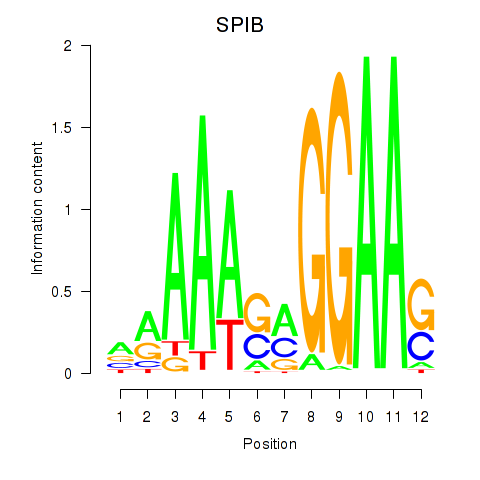
Results for SPIB
Z-value: 0.82
Transcription factors associated with SPIB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SPIB
|
ENSG00000269404.2 | Spi-B transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SPIB | hg19_v2_chr19_+_50922187_50922215 | 0.90 | 1.0e-01 | Click! |
Activity profile of SPIB motif
Sorted Z-values of SPIB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_108921977 | 0.54 |
ENST00000430559.1
ENST00000375887.4 |
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr8_-_131028782 | 0.53 |
ENST00000519020.1
|
FAM49B
|
family with sequence similarity 49, member B |
| chr14_-_80678512 | 0.46 |
ENST00000553968.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chr14_+_38065052 | 0.40 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr3_-_114035026 | 0.39 |
ENST00000570269.1
|
RP11-553L6.5
|
RP11-553L6.5 |
| chr22_-_37880543 | 0.38 |
ENST00000442496.1
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr15_+_91427642 | 0.37 |
ENST00000328850.3
ENST00000414248.2 |
FES
|
feline sarcoma oncogene |
| chr15_+_43885799 | 0.36 |
ENST00000449946.1
ENST00000417289.1 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr5_-_156390230 | 0.35 |
ENST00000407087.3
ENST00000274532.2 |
TIMD4
|
T-cell immunoglobulin and mucin domain containing 4 |
| chr17_-_27467418 | 0.34 |
ENST00000528564.1
|
MYO18A
|
myosin XVIIIA |
| chr19_+_50919056 | 0.33 |
ENST00000599632.1
|
CTD-2545M3.6
|
CTD-2545M3.6 |
| chr5_-_64858944 | 0.32 |
ENST00000508421.1
ENST00000510693.1 ENST00000514814.1 ENST00000515497.1 ENST00000396679.1 |
CENPK
|
centromere protein K |
| chr8_+_74903580 | 0.31 |
ENST00000284818.2
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr19_-_42192189 | 0.29 |
ENST00000401731.1
ENST00000338196.4 ENST00000006724.3 |
CEACAM7
|
carcinoembryonic antigen-related cell adhesion molecule 7 |
| chr16_+_54964740 | 0.29 |
ENST00000394636.4
|
IRX5
|
iroquois homeobox 5 |
| chr1_-_143913143 | 0.27 |
ENST00000400889.1
|
FAM72D
|
family with sequence similarity 72, member D |
| chr3_-_121379739 | 0.27 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr16_+_12059091 | 0.26 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr17_+_28443819 | 0.26 |
ENST00000479218.2
|
NSRP1
|
nuclear speckle splicing regulatory protein 1 |
| chr9_+_104296163 | 0.25 |
ENST00000374819.2
ENST00000479306.1 |
RNF20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chr20_-_62710832 | 0.25 |
ENST00000395042.1
|
RGS19
|
regulator of G-protein signaling 19 |
| chr19_+_42381337 | 0.25 |
ENST00000597454.1
ENST00000444740.2 |
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr3_+_28390637 | 0.24 |
ENST00000420223.1
ENST00000383768.2 |
ZCWPW2
|
zinc finger, CW type with PWWP domain 2 |
| chr6_+_49431073 | 0.24 |
ENST00000335783.3
|
CENPQ
|
centromere protein Q |
| chr5_-_142783175 | 0.23 |
ENST00000231509.3
ENST00000394464.2 |
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr15_-_65809991 | 0.23 |
ENST00000559526.1
ENST00000358939.4 ENST00000560665.1 ENST00000321118.7 ENST00000339244.5 ENST00000300141.6 |
DPP8
|
dipeptidyl-peptidase 8 |
| chr12_+_122018697 | 0.23 |
ENST00000541574.1
|
RP13-941N14.1
|
RP13-941N14.1 |
| chr20_-_62711259 | 0.23 |
ENST00000332298.5
|
RGS19
|
regulator of G-protein signaling 19 |
| chr10_+_106028923 | 0.23 |
ENST00000338595.2
|
GSTO2
|
glutathione S-transferase omega 2 |
| chr2_+_64751433 | 0.23 |
ENST00000238856.4
ENST00000422803.1 ENST00000238855.7 |
AFTPH
|
aftiphilin |
| chr3_+_184530173 | 0.22 |
ENST00000453056.1
|
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr6_-_49430886 | 0.22 |
ENST00000274813.3
|
MUT
|
methylmalonyl CoA mutase |
| chr5_-_142782862 | 0.22 |
ENST00000415690.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr3_+_98482175 | 0.22 |
ENST00000485391.1
ENST00000492254.1 |
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr20_-_46415297 | 0.22 |
ENST00000467815.1
ENST00000359930.4 |
SULF2
|
sulfatase 2 |
| chr5_+_102455968 | 0.22 |
ENST00000358359.3
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr19_-_48867291 | 0.21 |
ENST00000435956.3
|
TMEM143
|
transmembrane protein 143 |
| chr3_+_16926441 | 0.21 |
ENST00000418129.2
ENST00000396755.2 |
PLCL2
|
phospholipase C-like 2 |
| chr20_-_46415341 | 0.21 |
ENST00000484875.1
ENST00000361612.4 |
SULF2
|
sulfatase 2 |
| chr4_-_681114 | 0.21 |
ENST00000503156.1
|
MFSD7
|
major facilitator superfamily domain containing 7 |
| chr1_+_104068312 | 0.21 |
ENST00000524631.1
ENST00000531883.1 ENST00000533099.1 ENST00000527062.1 |
RNPC3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr1_+_164528616 | 0.21 |
ENST00000340699.3
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr3_-_143567262 | 0.21 |
ENST00000474151.1
ENST00000316549.6 |
SLC9A9
|
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chr20_+_62492566 | 0.21 |
ENST00000369916.3
|
ABHD16B
|
abhydrolase domain containing 16B |
| chr6_+_42952237 | 0.20 |
ENST00000485511.1
ENST00000394110.3 ENST00000472118.1 ENST00000461010.1 |
PPP2R5D
|
protein phosphatase 2, regulatory subunit B', delta |
| chr1_+_161475208 | 0.20 |
ENST00000367972.4
ENST00000271450.6 |
FCGR2A
|
Fc fragment of IgG, low affinity IIa, receptor (CD32) |
| chr19_-_48867171 | 0.20 |
ENST00000377431.2
ENST00000436660.2 ENST00000541566.1 |
TMEM143
|
transmembrane protein 143 |
| chr2_-_152118276 | 0.20 |
ENST00000409092.1
|
RBM43
|
RNA binding motif protein 43 |
| chr20_-_35274548 | 0.19 |
ENST00000262866.4
|
SLA2
|
Src-like-adaptor 2 |
| chr1_+_32757718 | 0.19 |
ENST00000428704.1
|
HDAC1
|
histone deacetylase 1 |
| chr19_-_42758040 | 0.19 |
ENST00000593944.1
|
ERF
|
Ets2 repressor factor |
| chr10_-_121302195 | 0.19 |
ENST00000369103.2
|
RGS10
|
regulator of G-protein signaling 10 |
| chr1_+_112016414 | 0.19 |
ENST00000343534.5
ENST00000369718.3 |
C1orf162
|
chromosome 1 open reading frame 162 |
| chr10_+_106028605 | 0.18 |
ENST00000450629.2
|
GSTO2
|
glutathione S-transferase omega 2 |
| chr8_-_125551278 | 0.18 |
ENST00000519232.1
ENST00000523888.1 ENST00000522810.1 ENST00000519548.1 ENST00000517678.1 ENST00000605953.1 ENST00000276692.6 |
TATDN1
|
TatD DNase domain containing 1 |
| chr16_+_57576584 | 0.18 |
ENST00000340339.4
|
GPR114
|
G protein-coupled receptor 114 |
| chr6_-_34524049 | 0.18 |
ENST00000374037.3
|
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chr15_-_65809581 | 0.18 |
ENST00000341861.5
|
DPP8
|
dipeptidyl-peptidase 8 |
| chr4_+_76871883 | 0.18 |
ENST00000599764.1
|
AC110615.1
|
Uncharacterized protein |
| chr17_-_43487741 | 0.18 |
ENST00000455881.1
|
ARHGAP27
|
Rho GTPase activating protein 27 |
| chr3_+_14474178 | 0.18 |
ENST00000452775.1
|
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter), member 6 |
| chr16_+_30960375 | 0.17 |
ENST00000318663.4
ENST00000566237.1 ENST00000562699.1 |
ORAI3
|
ORAI calcium release-activated calcium modulator 3 |
| chr11_-_46722117 | 0.17 |
ENST00000311956.4
|
ARHGAP1
|
Rho GTPase activating protein 1 |
| chr1_-_159832438 | 0.16 |
ENST00000368100.1
|
VSIG8
|
V-set and immunoglobulin domain containing 8 |
| chrX_+_24072833 | 0.16 |
ENST00000253039.4
|
EIF2S3
|
eukaryotic translation initiation factor 2, subunit 3 gamma, 52kDa |
| chr12_+_54447637 | 0.16 |
ENST00000609810.1
ENST00000430889.2 |
HOXC4
HOXC4
|
homeobox C4 Homeobox protein Hox-C4 |
| chr5_+_64859066 | 0.16 |
ENST00000261308.5
ENST00000535264.1 ENST00000538977.1 |
PPWD1
|
peptidylprolyl isomerase domain and WD repeat containing 1 |
| chr2_-_8723918 | 0.16 |
ENST00000454224.1
|
AC011747.4
|
AC011747.4 |
| chr11_-_60720002 | 0.16 |
ENST00000538739.1
|
SLC15A3
|
solute carrier family 15 (oligopeptide transporter), member 3 |
| chr1_+_206138884 | 0.16 |
ENST00000341209.5
ENST00000607379.1 |
FAM72A
|
family with sequence similarity 72, member A |
| chr11_-_10920838 | 0.16 |
ENST00000503469.2
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr2_-_180871780 | 0.16 |
ENST00000410053.3
ENST00000295749.6 ENST00000404136.2 |
CWC22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr14_+_75536335 | 0.16 |
ENST00000554763.1
ENST00000439583.2 ENST00000526130.1 ENST00000525046.1 |
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr16_-_30381580 | 0.16 |
ENST00000409939.3
|
TBC1D10B
|
TBC1 domain family, member 10B |
| chr4_+_95129061 | 0.16 |
ENST00000354268.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr10_-_977564 | 0.16 |
ENST00000406525.2
|
LARP4B
|
La ribonucleoprotein domain family, member 4B |
| chr11_+_3875757 | 0.15 |
ENST00000525403.1
|
STIM1
|
stromal interaction molecule 1 |
| chr15_+_91427691 | 0.15 |
ENST00000559355.1
ENST00000394302.1 |
FES
|
feline sarcoma oncogene |
| chr15_-_65810042 | 0.15 |
ENST00000321147.6
|
DPP8
|
dipeptidyl-peptidase 8 |
| chr6_-_144385698 | 0.15 |
ENST00000444202.1
ENST00000437412.1 |
PLAGL1
|
pleiomorphic adenoma gene-like 1 |
| chr5_+_92919043 | 0.15 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr7_-_122526499 | 0.15 |
ENST00000412584.2
|
CADPS2
|
Ca++-dependent secretion activator 2 |
| chr3_-_186080012 | 0.14 |
ENST00000544847.1
ENST00000265022.3 |
DGKG
|
diacylglycerol kinase, gamma 90kDa |
| chr17_-_4642429 | 0.14 |
ENST00000573123.1
|
CXCL16
|
chemokine (C-X-C motif) ligand 16 |
| chr9_-_113800981 | 0.14 |
ENST00000538760.1
|
LPAR1
|
lysophosphatidic acid receptor 1 |
| chr3_-_20227619 | 0.13 |
ENST00000425061.1
ENST00000443724.1 ENST00000421451.1 ENST00000452020.1 ENST00000417364.1 ENST00000306698.2 ENST00000419233.2 ENST00000263753.4 ENST00000383774.1 ENST00000437051.1 ENST00000412868.1 ENST00000429446.3 ENST00000442720.1 |
SGOL1
|
shugoshin-like 1 (S. pombe) |
| chr17_+_76356516 | 0.13 |
ENST00000592569.1
|
RP11-806H10.4
|
RP11-806H10.4 |
| chr4_-_47916543 | 0.13 |
ENST00000507489.1
|
NFXL1
|
nuclear transcription factor, X-box binding-like 1 |
| chr8_-_101734308 | 0.13 |
ENST00000519004.1
ENST00000519363.1 ENST00000520142.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr12_-_15114603 | 0.13 |
ENST00000228945.4
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr2_+_233925064 | 0.13 |
ENST00000359570.5
ENST00000538935.1 |
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa |
| chr9_-_95640218 | 0.13 |
ENST00000395506.3
ENST00000375495.3 ENST00000332591.6 |
ZNF484
|
zinc finger protein 484 |
| chr12_-_53901266 | 0.13 |
ENST00000609999.1
ENST00000267017.3 |
NPFF
|
neuropeptide FF-amide peptide precursor |
| chr14_+_102276192 | 0.13 |
ENST00000557714.1
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr9_-_14693417 | 0.13 |
ENST00000380916.4
|
ZDHHC21
|
zinc finger, DHHC-type containing 21 |
| chr6_+_22221010 | 0.13 |
ENST00000567753.1
|
RP11-524C21.2
|
RP11-524C21.2 |
| chr1_+_173991633 | 0.13 |
ENST00000424181.1
|
RP11-160H22.3
|
RP11-160H22.3 |
| chr2_+_135011731 | 0.13 |
ENST00000281923.2
|
MGAT5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr20_+_20033158 | 0.13 |
ENST00000340348.6
ENST00000377309.2 ENST00000389656.3 ENST00000377306.1 ENST00000245957.5 ENST00000377303.2 ENST00000475466.1 |
C20orf26
|
chromosome 20 open reading frame 26 |
| chr1_-_43282906 | 0.12 |
ENST00000372521.4
|
CCDC23
|
coiled-coil domain containing 23 |
| chr16_+_70695570 | 0.12 |
ENST00000597002.1
|
FLJ00418
|
FLJ00418 |
| chr3_-_20227720 | 0.12 |
ENST00000412997.1
|
SGOL1
|
shugoshin-like 1 (S. pombe) |
| chr14_-_74416829 | 0.12 |
ENST00000534936.1
|
FAM161B
|
family with sequence similarity 161, member B |
| chr19_-_15344243 | 0.12 |
ENST00000602233.1
|
EPHX3
|
epoxide hydrolase 3 |
| chr1_+_14075865 | 0.12 |
ENST00000413440.1
|
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr15_-_45422056 | 0.12 |
ENST00000267803.4
ENST00000559014.1 ENST00000558851.1 ENST00000559988.1 ENST00000558996.1 ENST00000558422.1 ENST00000559226.1 ENST00000558326.1 ENST00000558377.1 ENST00000559644.1 |
DUOXA1
|
dual oxidase maturation factor 1 |
| chr15_+_59279851 | 0.12 |
ENST00000348370.4
ENST00000434298.1 ENST00000559160.1 |
RNF111
|
ring finger protein 111 |
| chr1_+_120839412 | 0.12 |
ENST00000355228.4
|
FAM72B
|
family with sequence similarity 72, member B |
| chr8_-_37707356 | 0.12 |
ENST00000520601.1
ENST00000521170.1 ENST00000220659.6 |
BRF2
|
BRF2, RNA polymerase III transcription initiation factor 50 kDa subunit |
| chr6_+_130339710 | 0.12 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr5_-_40835303 | 0.12 |
ENST00000509877.1
ENST00000508493.1 ENST00000274242.5 |
RPL37
|
ribosomal protein L37 |
| chr3_-_9291063 | 0.11 |
ENST00000383836.3
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr6_-_34524093 | 0.11 |
ENST00000544425.1
|
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chr8_+_104426942 | 0.11 |
ENST00000297579.5
|
DCAF13
|
DDB1 and CUL4 associated factor 13 |
| chr1_+_179851823 | 0.11 |
ENST00000435319.4
|
TOR1AIP1
|
torsin A interacting protein 1 |
| chr2_-_153573887 | 0.11 |
ENST00000493468.2
ENST00000545856.1 |
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr3_+_98482159 | 0.11 |
ENST00000468553.1
|
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr4_-_140005443 | 0.11 |
ENST00000510408.1
ENST00000420916.2 ENST00000358635.3 |
ELF2
|
E74-like factor 2 (ets domain transcription factor) |
| chr5_+_102455853 | 0.11 |
ENST00000515845.1
ENST00000321521.9 ENST00000507921.1 |
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr11_-_1606513 | 0.11 |
ENST00000382171.2
|
KRTAP5-1
|
keratin associated protein 5-1 |
| chr20_-_20033052 | 0.11 |
ENST00000536226.1
|
CRNKL1
|
crooked neck pre-mRNA splicing factor 1 |
| chr17_-_43487780 | 0.11 |
ENST00000532038.1
ENST00000528677.1 |
ARHGAP27
|
Rho GTPase activating protein 27 |
| chr12_+_32638897 | 0.11 |
ENST00000531134.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr3_+_184529929 | 0.11 |
ENST00000287546.4
ENST00000437079.3 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr1_-_220445757 | 0.11 |
ENST00000358951.2
|
RAB3GAP2
|
RAB3 GTPase activating protein subunit 2 (non-catalytic) |
| chr19_+_10959043 | 0.11 |
ENST00000397820.4
|
C19orf38
|
chromosome 19 open reading frame 38 |
| chr5_-_10249990 | 0.11 |
ENST00000511437.1
ENST00000280330.8 ENST00000510047.1 |
FAM173B
|
family with sequence similarity 173, member B |
| chr5_+_35856951 | 0.11 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr6_+_31126291 | 0.11 |
ENST00000376257.3
ENST00000376255.4 |
TCF19
|
transcription factor 19 |
| chr17_-_29641104 | 0.10 |
ENST00000577894.1
ENST00000330927.4 |
EVI2B
|
ecotropic viral integration site 2B |
| chr15_+_42651691 | 0.10 |
ENST00000357568.3
ENST00000349748.3 ENST00000318023.7 ENST00000397163.3 |
CAPN3
|
calpain 3, (p94) |
| chr19_-_18391708 | 0.10 |
ENST00000600972.1
|
JUND
|
jun D proto-oncogene |
| chr3_-_28390581 | 0.10 |
ENST00000479665.1
|
AZI2
|
5-azacytidine induced 2 |
| chr19_-_9420249 | 0.10 |
ENST00000591998.1
|
ZNF699
|
zinc finger protein 699 |
| chr16_-_30537839 | 0.10 |
ENST00000380412.5
|
ZNF768
|
zinc finger protein 768 |
| chr1_+_209941827 | 0.10 |
ENST00000367023.1
|
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr1_-_20446020 | 0.10 |
ENST00000375105.3
|
PLA2G2D
|
phospholipase A2, group IID |
| chr19_+_51728316 | 0.10 |
ENST00000436584.2
ENST00000421133.2 ENST00000391796.3 ENST00000262262.4 |
CD33
|
CD33 molecule |
| chr12_-_15114492 | 0.10 |
ENST00000541546.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr17_+_72667239 | 0.10 |
ENST00000402449.4
|
RAB37
|
RAB37, member RAS oncogene family |
| chr9_-_125675576 | 0.10 |
ENST00000373659.3
|
ZBTB6
|
zinc finger and BTB domain containing 6 |
| chr12_-_15114191 | 0.10 |
ENST00000541380.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr12_-_8218997 | 0.10 |
ENST00000307637.4
|
C3AR1
|
complement component 3a receptor 1 |
| chr1_+_206138457 | 0.10 |
ENST00000367128.3
ENST00000431655.2 |
FAM72A
|
family with sequence similarity 72, member A |
| chr7_-_37488834 | 0.09 |
ENST00000310758.4
|
ELMO1
|
engulfment and cell motility 1 |
| chr1_+_120839005 | 0.09 |
ENST00000369390.3
ENST00000452190.1 |
FAM72B
|
family with sequence similarity 72, member B |
| chr12_-_110939870 | 0.09 |
ENST00000447578.2
ENST00000546588.1 ENST00000360579.7 ENST00000549970.1 ENST00000549578.1 |
VPS29
|
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr5_+_65892174 | 0.09 |
ENST00000404260.3
ENST00000403625.2 ENST00000406374.1 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr19_-_40790729 | 0.09 |
ENST00000423127.1
ENST00000583859.1 ENST00000456441.1 ENST00000452077.1 |
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr3_-_116163830 | 0.09 |
ENST00000333617.4
|
LSAMP
|
limbic system-associated membrane protein |
| chr11_-_61124711 | 0.09 |
ENST00000536915.1
|
CYB561A3
|
cytochrome b561 family, member A3 |
| chr9_-_130541017 | 0.09 |
ENST00000314830.8
|
SH2D3C
|
SH2 domain containing 3C |
| chr1_+_159272111 | 0.09 |
ENST00000368114.1
|
FCER1A
|
Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide |
| chr19_-_4559814 | 0.09 |
ENST00000586582.1
|
SEMA6B
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
| chr2_-_113993020 | 0.09 |
ENST00000465084.1
|
PAX8
|
paired box 8 |
| chr1_+_151171012 | 0.09 |
ENST00000349792.5
ENST00000409426.1 ENST00000441902.2 ENST00000368890.4 ENST00000424999.1 ENST00000368888.4 |
PIP5K1A
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha |
| chr3_-_155462808 | 0.09 |
ENST00000460012.1
|
PLCH1
|
phospholipase C, eta 1 |
| chr7_-_150038704 | 0.09 |
ENST00000466675.1
ENST00000482669.1 ENST00000467793.1 ENST00000223271.3 |
RARRES2
|
retinoic acid receptor responder (tazarotene induced) 2 |
| chr1_+_110158726 | 0.08 |
ENST00000531734.1
|
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr6_+_31554779 | 0.08 |
ENST00000376090.2
|
LST1
|
leukocyte specific transcript 1 |
| chr7_+_139528952 | 0.08 |
ENST00000416849.2
ENST00000436047.2 ENST00000414508.2 ENST00000448866.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr3_+_184529948 | 0.08 |
ENST00000436792.2
ENST00000446204.2 ENST00000422105.1 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr19_-_42192096 | 0.08 |
ENST00000602225.1
|
CEACAM7
|
carcinoembryonic antigen-related cell adhesion molecule 7 |
| chr19_+_1383890 | 0.08 |
ENST00000539480.1
ENST00000313408.7 ENST00000414651.2 |
NDUFS7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7, 20kDa (NADH-coenzyme Q reductase) |
| chr1_+_15783222 | 0.08 |
ENST00000359621.4
|
CELA2A
|
chymotrypsin-like elastase family, member 2A |
| chr14_+_102276132 | 0.08 |
ENST00000350249.3
ENST00000557621.1 ENST00000556946.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr4_-_102268708 | 0.08 |
ENST00000525819.1
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr12_-_120663792 | 0.08 |
ENST00000546532.1
ENST00000548912.1 |
PXN
|
paxillin |
| chr4_-_146859787 | 0.08 |
ENST00000508784.1
|
ZNF827
|
zinc finger protein 827 |
| chr15_+_43985084 | 0.08 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr4_+_86525299 | 0.08 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr8_+_27632083 | 0.08 |
ENST00000519637.1
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr12_+_49717019 | 0.08 |
ENST00000549275.1
ENST00000551245.1 ENST00000380327.5 ENST00000548311.1 ENST00000550346.1 ENST00000550709.1 ENST00000549534.1 ENST00000257909.3 |
TROAP
|
trophinin associated protein |
| chrX_+_24073048 | 0.08 |
ENST00000423068.1
|
EIF2S3
|
eukaryotic translation initiation factor 2, subunit 3 gamma, 52kDa |
| chr14_-_50319482 | 0.08 |
ENST00000546046.1
ENST00000555970.1 ENST00000554626.1 ENST00000545773.1 ENST00000556672.1 |
NEMF
|
nuclear export mediator factor |
| chr17_-_56492989 | 0.08 |
ENST00000583753.1
|
RNF43
|
ring finger protein 43 |
| chr15_+_43885252 | 0.08 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr1_-_175161890 | 0.07 |
ENST00000545251.2
ENST00000423313.1 |
KIAA0040
|
KIAA0040 |
| chr1_+_161551101 | 0.07 |
ENST00000367962.4
ENST00000367960.5 ENST00000403078.3 ENST00000428605.2 |
FCGR2B
|
Fc fragment of IgG, low affinity IIb, receptor (CD32) |
| chr14_+_96968802 | 0.07 |
ENST00000556619.1
ENST00000392990.2 |
PAPOLA
|
poly(A) polymerase alpha |
| chr10_+_18429671 | 0.07 |
ENST00000282343.8
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr14_+_75536280 | 0.07 |
ENST00000238686.8
|
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr3_-_195603566 | 0.07 |
ENST00000424563.1
ENST00000411741.1 |
TNK2
|
tyrosine kinase, non-receptor, 2 |
| chrX_+_24167746 | 0.07 |
ENST00000428571.1
ENST00000539115.1 |
ZFX
|
zinc finger protein, X-linked |
| chr11_-_18656028 | 0.07 |
ENST00000336349.5
|
SPTY2D1
|
SPT2, Suppressor of Ty, domain containing 1 (S. cerevisiae) |
| chr14_+_102276209 | 0.07 |
ENST00000445439.3
ENST00000334743.5 ENST00000557095.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr6_-_110012380 | 0.07 |
ENST00000424296.2
ENST00000341338.6 ENST00000368948.2 ENST00000285397.5 |
AK9
|
adenylate kinase 9 |
| chr16_+_31213206 | 0.07 |
ENST00000561916.2
|
C16orf98
|
chromosome 16 open reading frame 98 |
| chr2_+_162087577 | 0.07 |
ENST00000439442.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr14_+_45431379 | 0.07 |
ENST00000361577.3
ENST00000361462.2 ENST00000382233.2 |
FAM179B
|
family with sequence similarity 179, member B |
| chr2_-_26101314 | 0.07 |
ENST00000336112.4
ENST00000272341.4 |
ASXL2
|
additional sex combs like 2 (Drosophila) |
| chr14_-_50319758 | 0.07 |
ENST00000298310.5
|
NEMF
|
nuclear export mediator factor |
| chr2_+_102758271 | 0.07 |
ENST00000428279.1
|
IL1R1
|
interleukin 1 receptor, type I |
| chr14_-_98444461 | 0.07 |
ENST00000499006.2
|
C14orf64
|
chromosome 14 open reading frame 64 |
| chr12_+_10124001 | 0.07 |
ENST00000396507.3
ENST00000304361.4 ENST00000434319.2 |
CLEC12A
|
C-type lectin domain family 12, member A |
| chr19_+_2476116 | 0.07 |
ENST00000215631.4
ENST00000587345.1 |
GADD45B
|
growth arrest and DNA-damage-inducible, beta |
| chr5_+_159848854 | 0.07 |
ENST00000517480.1
ENST00000520452.1 ENST00000393964.1 |
PTTG1
|
pituitary tumor-transforming 1 |
| chr3_-_176914238 | 0.07 |
ENST00000430069.1
ENST00000428970.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr1_-_45956822 | 0.07 |
ENST00000372086.3
ENST00000341771.6 |
TESK2
|
testis-specific kinase 2 |
| chr2_-_152830479 | 0.07 |
ENST00000360283.6
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
Network of associatons between targets according to the STRING database.
First level regulatory network of SPIB
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.1 | 0.4 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.3 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 0.3 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.1 | 0.5 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.1 | 0.2 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 0.3 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.1 | 0.3 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 | 0.3 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.0 | 0.2 | GO:0016068 | regulation of type I hypersensitivity(GO:0001810) type I hypersensitivity(GO:0016068) |
| 0.0 | 0.1 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.2 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.5 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.3 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.0 | 0.4 | GO:0090487 | toxin catabolic process(GO:0009407) secondary metabolite catabolic process(GO:0090487) |
| 0.0 | 0.1 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.0 | 0.7 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.2 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.1 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 0.0 | 0.2 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.5 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.1 | GO:0045404 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 0.1 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.3 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.2 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.0 | GO:0002461 | tolerance induction dependent upon immune response(GO:0002461) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0061566 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.0 | 0.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.1 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.1 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.1 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.0 | 0.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.0 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.0 | 0.0 | GO:0097065 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.3 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.0 | GO:0045399 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.1 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.0 | 0.6 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0042335 | cuticle development(GO:0042335) |
| 0.0 | 0.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.0 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.3 | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling(GO:0014066) |
| 0.0 | 0.2 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.3 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.4 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.2 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.1 | GO:0036457 | keratohyalin granule(GO:0036457) |
| 0.0 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.1 | 0.4 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.3 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.2 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 0.4 | GO:0045174 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.1 | 0.5 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.3 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.1 | 0.5 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.3 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.3 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.0 | 0.1 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.0 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.1 | GO:0052811 | 1-phosphatidylinositol-3-phosphate 4-kinase activity(GO:0052811) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.1 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.0 | 0.1 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.2 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.5 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.1 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.0 | 0.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |