Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
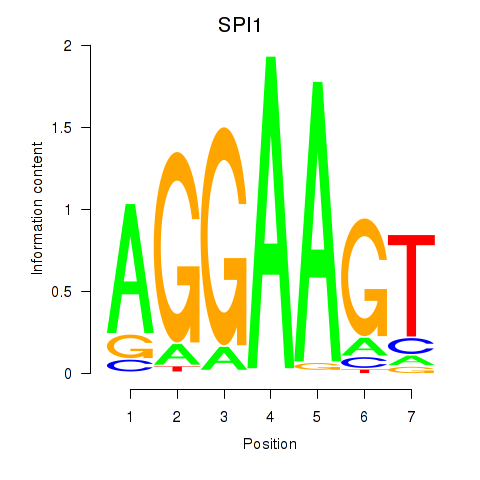
Results for SPI1
Z-value: 0.34
Transcription factors associated with SPI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SPI1
|
ENSG00000066336.7 | Spi-1 proto-oncogene |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SPI1 | hg19_v2_chr11_-_47399942_47399961 | 0.82 | 1.8e-01 | Click! |
Activity profile of SPI1 motif
Sorted Z-values of SPI1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_119013185 | 0.11 |
ENST00000264245.4
|
ARHGAP31
|
Rho GTPase activating protein 31 |
| chr5_-_175388120 | 0.11 |
ENST00000515016.1
|
THOC3
|
THO complex 3 |
| chr6_+_139349903 | 0.10 |
ENST00000461027.1
|
ABRACL
|
ABRA C-terminal like |
| chr17_+_7461613 | 0.09 |
ENST00000438470.1
ENST00000436057.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr6_+_26156551 | 0.09 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr19_+_36393367 | 0.09 |
ENST00000246551.4
|
HCST
|
hematopoietic cell signal transducer |
| chr1_-_101360205 | 0.09 |
ENST00000450240.1
|
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr16_-_4039001 | 0.08 |
ENST00000576936.1
|
ADCY9
|
adenylate cyclase 9 |
| chr21_+_17961006 | 0.08 |
ENST00000602323.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr16_-_67260691 | 0.08 |
ENST00000447579.1
ENST00000393992.1 ENST00000424285.1 |
LRRC29
|
leucine rich repeat containing 29 |
| chr7_-_37382683 | 0.08 |
ENST00000455879.1
|
ELMO1
|
engulfment and cell motility 1 |
| chr16_-_30204987 | 0.07 |
ENST00000569282.1
ENST00000567436.1 |
BOLA2B
|
bolA family member 2B |
| chr16_-_67260901 | 0.07 |
ENST00000341546.3
ENST00000409509.1 ENST00000433915.1 ENST00000454102.2 |
LRRC29
AC040160.1
|
leucine rich repeat containing 29 Uncharacterized protein; cDNA FLJ57407, weakly similar to Mus musculus leucine rich repeat containing 29 (Lrrc29), mRNA |
| chr22_+_37257015 | 0.07 |
ENST00000447071.1
ENST00000248899.6 ENST00000397147.4 |
NCF4
|
neutrophil cytosolic factor 4, 40kDa |
| chr7_+_114055052 | 0.07 |
ENST00000462331.1
ENST00000408937.3 ENST00000403559.4 ENST00000350908.4 ENST00000393498.2 ENST00000393495.3 ENST00000378237.3 ENST00000393489.3 |
FOXP2
|
forkhead box P2 |
| chr12_+_14572070 | 0.07 |
ENST00000545769.1
ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr2_+_87755054 | 0.07 |
ENST00000423846.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr8_+_74903580 | 0.07 |
ENST00000284818.2
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr16_-_30393752 | 0.07 |
ENST00000566517.1
ENST00000605106.1 |
SEPT1
SEPT1
|
septin 1 Uncharacterized protein |
| chr7_+_106505696 | 0.07 |
ENST00000440650.2
ENST00000496166.1 ENST00000473541.1 |
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr1_-_151254362 | 0.06 |
ENST00000447795.2
|
RP11-126K1.2
|
Uncharacterized protein |
| chr19_+_41281282 | 0.06 |
ENST00000263369.3
|
MIA
|
melanoma inhibitory activity |
| chr17_+_7461849 | 0.06 |
ENST00000338784.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr2_-_97534312 | 0.06 |
ENST00000442264.1
|
SEMA4C
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr11_+_18154059 | 0.06 |
ENST00000531264.1
|
MRGPRX3
|
MAS-related GPR, member X3 |
| chr15_-_33360342 | 0.06 |
ENST00000558197.1
|
FMN1
|
formin 1 |
| chr9_+_120466650 | 0.06 |
ENST00000355622.6
|
TLR4
|
toll-like receptor 4 |
| chr1_-_179851611 | 0.06 |
ENST00000610272.1
|
RP11-533E19.7
|
RP11-533E19.7 |
| chr19_-_39303576 | 0.06 |
ENST00000594209.1
|
LGALS4
|
lectin, galactoside-binding, soluble, 4 |
| chr9_+_139839711 | 0.05 |
ENST00000224181.3
|
C8G
|
complement component 8, gamma polypeptide |
| chr9_+_108424738 | 0.05 |
ENST00000334077.3
|
TAL2
|
T-cell acute lymphocytic leukemia 2 |
| chr2_-_32390801 | 0.05 |
ENST00000608489.1
|
RP11-563N4.1
|
RP11-563N4.1 |
| chr14_-_76447494 | 0.05 |
ENST00000238682.3
|
TGFB3
|
transforming growth factor, beta 3 |
| chr11_+_36397915 | 0.05 |
ENST00000526682.1
ENST00000530252.1 |
PRR5L
|
proline rich 5 like |
| chr3_-_11762202 | 0.05 |
ENST00000445411.1
ENST00000404339.1 ENST00000273038.3 |
VGLL4
|
vestigial like 4 (Drosophila) |
| chr20_+_44746939 | 0.05 |
ENST00000372276.3
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5 |
| chr2_-_198540751 | 0.05 |
ENST00000429081.1
|
RFTN2
|
raftlin family member 2 |
| chr16_+_67063262 | 0.05 |
ENST00000565389.1
|
CBFB
|
core-binding factor, beta subunit |
| chr11_-_134095335 | 0.05 |
ENST00000534227.1
ENST00000532445.1 |
NCAPD3
|
non-SMC condensin II complex, subunit D3 |
| chr6_-_34524093 | 0.05 |
ENST00000544425.1
|
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chr5_-_175388327 | 0.05 |
ENST00000432305.2
ENST00000505969.1 |
THOC3
|
THO complex 3 |
| chr16_+_72142195 | 0.05 |
ENST00000563819.1
ENST00000567142.2 |
DHX38
|
DEAH (Asp-Glu-Ala-His) box polypeptide 38 |
| chr11_-_75141206 | 0.05 |
ENST00000376292.4
|
KLHL35
|
kelch-like family member 35 |
| chr4_-_156297919 | 0.05 |
ENST00000450097.1
|
MAP9
|
microtubule-associated protein 9 |
| chr16_-_85969774 | 0.05 |
ENST00000598933.1
|
RP11-542M13.3
|
RP11-542M13.3 |
| chr10_-_128110441 | 0.05 |
ENST00000456514.1
|
LINC00601
|
long intergenic non-protein coding RNA 601 |
| chr11_+_76813886 | 0.05 |
ENST00000529803.1
|
OMP
|
olfactory marker protein |
| chr19_+_17970693 | 0.05 |
ENST00000600147.1
ENST00000599898.1 |
RPL18A
|
ribosomal protein L18a |
| chr11_+_66824303 | 0.05 |
ENST00000533360.1
|
RHOD
|
ras homolog family member D |
| chr17_+_7461781 | 0.05 |
ENST00000349228.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr5_+_35856951 | 0.05 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr7_+_106505912 | 0.05 |
ENST00000359195.3
|
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr18_+_22040620 | 0.05 |
ENST00000426880.2
|
HRH4
|
histamine receptor H4 |
| chr5_+_61874562 | 0.05 |
ENST00000334994.5
ENST00000409534.1 |
LRRC70
IPO11
|
leucine rich repeat containing 70 importin 11 |
| chr17_+_6918354 | 0.05 |
ENST00000552775.1
|
C17orf49
|
chromosome 17 open reading frame 49 |
| chr17_+_7461580 | 0.05 |
ENST00000483039.1
ENST00000396542.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr5_+_140474181 | 0.05 |
ENST00000194155.4
|
PCDHB2
|
protocadherin beta 2 |
| chr20_+_44746885 | 0.05 |
ENST00000372285.3
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5 |
| chr12_-_118796971 | 0.05 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3 |
| chr1_-_154909329 | 0.05 |
ENST00000368467.3
|
PMVK
|
phosphomevalonate kinase |
| chr8_+_99076750 | 0.05 |
ENST00000545282.1
|
C8orf47
|
chromosome 8 open reading frame 47 |
| chr17_-_7760779 | 0.05 |
ENST00000335155.5
ENST00000575071.1 |
LSMD1
|
LSM domain containing 1 |
| chr11_-_72432950 | 0.05 |
ENST00000426523.1
ENST00000429686.1 |
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr17_-_38256973 | 0.05 |
ENST00000246672.3
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1 |
| chr18_+_21594384 | 0.05 |
ENST00000584250.1
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr12_+_7055767 | 0.05 |
ENST00000447931.2
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr16_-_30394143 | 0.05 |
ENST00000321367.3
ENST00000571393.1 |
SEPT1
|
septin 1 |
| chr16_-_279405 | 0.05 |
ENST00000430864.1
ENST00000293872.8 ENST00000337351.4 ENST00000397783.1 |
LUC7L
|
LUC7-like (S. cerevisiae) |
| chr4_+_86396265 | 0.05 |
ENST00000395184.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr15_-_64385981 | 0.05 |
ENST00000557835.1
ENST00000380290.3 ENST00000559950.1 |
FAM96A
|
family with sequence similarity 96, member A |
| chr17_+_18218587 | 0.05 |
ENST00000406438.3
|
SMCR8
|
Smith-Magenis syndrome chromosome region, candidate 8 |
| chr3_+_11267691 | 0.05 |
ENST00000413416.1
|
HRH1
|
histamine receptor H1 |
| chr18_-_60985914 | 0.04 |
ENST00000589955.1
|
BCL2
|
B-cell CLL/lymphoma 2 |
| chr6_+_31620191 | 0.04 |
ENST00000375918.2
ENST00000375920.4 |
APOM
|
apolipoprotein M |
| chr17_+_45908974 | 0.04 |
ENST00000269025.4
|
LRRC46
|
leucine rich repeat containing 46 |
| chr6_-_34524049 | 0.04 |
ENST00000374037.3
|
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chrX_-_153979315 | 0.04 |
ENST00000369575.3
ENST00000369568.4 ENST00000424127.2 |
GAB3
|
GRB2-associated binding protein 3 |
| chr20_-_31124186 | 0.04 |
ENST00000375678.3
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr3_+_119316721 | 0.04 |
ENST00000488919.1
ENST00000495992.1 |
PLA1A
|
phospholipase A1 member A |
| chr17_-_28661065 | 0.04 |
ENST00000328886.4
ENST00000538566.2 |
TMIGD1
|
transmembrane and immunoglobulin domain containing 1 |
| chr1_-_205912577 | 0.04 |
ENST00000367135.3
ENST00000367134.2 |
SLC26A9
|
solute carrier family 26 (anion exchanger), member 9 |
| chr6_-_127840048 | 0.04 |
ENST00000467753.1
|
SOGA3
|
SOGA family member 3 |
| chr11_-_102826434 | 0.04 |
ENST00000340273.4
ENST00000260302.3 |
MMP13
|
matrix metallopeptidase 13 (collagenase 3) |
| chr11_+_65408273 | 0.04 |
ENST00000394227.3
|
SIPA1
|
signal-induced proliferation-associated 1 |
| chr12_+_7055631 | 0.04 |
ENST00000543115.1
ENST00000399448.1 |
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr1_+_159272111 | 0.04 |
ENST00000368114.1
|
FCER1A
|
Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide |
| chr12_+_117176090 | 0.04 |
ENST00000257575.4
ENST00000407967.3 ENST00000392549.2 |
RNFT2
|
ring finger protein, transmembrane 2 |
| chr17_+_58677539 | 0.04 |
ENST00000305921.3
|
PPM1D
|
protein phosphatase, Mg2+/Mn2+ dependent, 1D |
| chr9_-_134145880 | 0.04 |
ENST00000372269.3
ENST00000464831.1 |
FAM78A
|
family with sequence similarity 78, member A |
| chr17_+_2264983 | 0.04 |
ENST00000574650.1
|
SGSM2
|
small G protein signaling modulator 2 |
| chr6_+_29555683 | 0.04 |
ENST00000383640.2
|
OR2H2
|
olfactory receptor, family 2, subfamily H, member 2 |
| chr19_+_17970677 | 0.04 |
ENST00000222247.5
|
RPL18A
|
ribosomal protein L18a |
| chr7_-_27219632 | 0.04 |
ENST00000470747.4
|
RP1-170O19.20
|
Uncharacterized protein |
| chr11_-_72433346 | 0.04 |
ENST00000334211.8
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr11_+_66824276 | 0.04 |
ENST00000308831.2
|
RHOD
|
ras homolog family member D |
| chr17_-_7761256 | 0.04 |
ENST00000575208.1
|
LSMD1
|
LSM domain containing 1 |
| chr6_-_33168391 | 0.04 |
ENST00000374685.4
ENST00000413614.2 ENST00000374680.3 |
RXRB
|
retinoid X receptor, beta |
| chr17_+_65027509 | 0.04 |
ENST00000375684.1
|
AC005544.1
|
Uncharacterized protein |
| chr14_+_52327350 | 0.04 |
ENST00000555472.1
ENST00000556766.1 |
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr12_+_117176113 | 0.04 |
ENST00000319176.7
|
RNFT2
|
ring finger protein, transmembrane 2 |
| chr7_-_37382360 | 0.04 |
ENST00000455119.1
|
ELMO1
|
engulfment and cell motility 1 |
| chr14_+_23025534 | 0.04 |
ENST00000557595.1
|
AE000662.92
|
Uncharacterized protein |
| chr17_-_7761172 | 0.04 |
ENST00000333775.5
ENST00000575771.1 |
LSMD1
|
LSM domain containing 1 |
| chr11_+_46366918 | 0.04 |
ENST00000528615.1
ENST00000395574.3 |
DGKZ
|
diacylglycerol kinase, zeta |
| chr11_-_31832785 | 0.04 |
ENST00000241001.8
|
PAX6
|
paired box 6 |
| chr11_-_31832581 | 0.04 |
ENST00000379111.2
|
PAX6
|
paired box 6 |
| chr6_+_31553978 | 0.04 |
ENST00000376096.1
ENST00000376099.1 ENST00000376110.3 |
LST1
|
leukocyte specific transcript 1 |
| chr3_+_48481658 | 0.04 |
ENST00000438607.2
|
TMA7
|
translation machinery associated 7 homolog (S. cerevisiae) |
| chr17_-_34079897 | 0.04 |
ENST00000254466.6
ENST00000587565.1 |
GAS2L2
|
growth arrest-specific 2 like 2 |
| chr11_+_46369077 | 0.04 |
ENST00000456247.2
ENST00000421244.2 ENST00000318201.8 |
DGKZ
|
diacylglycerol kinase, zeta |
| chr16_-_30125177 | 0.04 |
ENST00000406256.3
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr9_+_95726243 | 0.04 |
ENST00000416701.2
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr2_-_69870835 | 0.04 |
ENST00000409085.4
ENST00000406297.3 |
AAK1
|
AP2 associated kinase 1 |
| chr2_-_154335300 | 0.04 |
ENST00000325926.3
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr9_+_116327326 | 0.04 |
ENST00000342620.5
|
RGS3
|
regulator of G-protein signaling 3 |
| chr4_+_87857538 | 0.04 |
ENST00000511442.1
|
AFF1
|
AF4/FMR2 family, member 1 |
| chr6_-_42418999 | 0.04 |
ENST00000340840.2
ENST00000354325.2 |
TRERF1
|
transcriptional regulating factor 1 |
| chr4_+_8201091 | 0.04 |
ENST00000382521.3
ENST00000245105.3 ENST00000457650.2 ENST00000539824.1 |
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1 |
| chrX_-_49056635 | 0.04 |
ENST00000472598.1
ENST00000538567.1 ENST00000479808.1 ENST00000263233.4 |
SYP
|
synaptophysin |
| chr11_-_64013288 | 0.04 |
ENST00000542235.1
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr17_+_37856299 | 0.04 |
ENST00000269571.5
|
ERBB2
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr2_+_39005336 | 0.04 |
ENST00000409566.1
|
GEMIN6
|
gem (nuclear organelle) associated protein 6 |
| chr9_+_120466610 | 0.04 |
ENST00000394487.4
|
TLR4
|
toll-like receptor 4 |
| chr12_-_118796910 | 0.04 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr12_+_12224331 | 0.04 |
ENST00000396367.1
ENST00000266434.4 ENST00000396369.1 |
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr4_-_48082192 | 0.04 |
ENST00000507351.1
|
TXK
|
TXK tyrosine kinase |
| chr12_-_123921256 | 0.04 |
ENST00000280571.8
|
RILPL2
|
Rab interacting lysosomal protein-like 2 |
| chr14_-_24701539 | 0.04 |
ENST00000534348.1
ENST00000524927.1 ENST00000250495.5 |
NEDD8-MDP1
NEDD8
|
NEDD8-MDP1 readthrough neural precursor cell expressed, developmentally down-regulated 8 |
| chr11_+_67219867 | 0.03 |
ENST00000438189.2
|
CABP4
|
calcium binding protein 4 |
| chr2_+_42104692 | 0.03 |
ENST00000398796.2
ENST00000442214.1 |
AC104654.1
|
AC104654.1 |
| chr17_+_37856253 | 0.03 |
ENST00000540147.1
ENST00000584450.1 |
ERBB2
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr17_-_74533734 | 0.03 |
ENST00000589342.1
|
CYGB
|
cytoglobin |
| chr11_-_130298888 | 0.03 |
ENST00000257359.6
|
ADAMTS8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8 |
| chr9_+_134000948 | 0.03 |
ENST00000359428.5
ENST00000411637.2 ENST00000451030.1 |
NUP214
|
nucleoporin 214kDa |
| chr19_+_14551066 | 0.03 |
ENST00000342216.4
|
PKN1
|
protein kinase N1 |
| chr1_+_203096831 | 0.03 |
ENST00000337894.4
|
ADORA1
|
adenosine A1 receptor |
| chr1_-_144994840 | 0.03 |
ENST00000369351.3
ENST00000369349.3 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr17_-_40134339 | 0.03 |
ENST00000587727.1
|
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chrX_-_153192211 | 0.03 |
ENST00000461052.1
ENST00000422091.1 |
ARHGAP4
|
Rho GTPase activating protein 4 |
| chr7_-_35293740 | 0.03 |
ENST00000408931.3
|
TBX20
|
T-box 20 |
| chr17_+_4618734 | 0.03 |
ENST00000571206.1
|
ARRB2
|
arrestin, beta 2 |
| chr22_-_30234218 | 0.03 |
ENST00000307790.3
ENST00000542393.1 ENST00000397771.2 |
ASCC2
|
activating signal cointegrator 1 complex subunit 2 |
| chr11_-_62358972 | 0.03 |
ENST00000278279.3
|
TUT1
|
terminal uridylyl transferase 1, U6 snRNA-specific |
| chr21_+_35107346 | 0.03 |
ENST00000456489.1
|
ITSN1
|
intersectin 1 (SH3 domain protein) |
| chr11_-_118889323 | 0.03 |
ENST00000527673.1
|
RPS25
|
ribosomal protein S25 |
| chr14_+_74004051 | 0.03 |
ENST00000557556.1
|
ACOT1
|
acyl-CoA thioesterase 1 |
| chr19_+_41313017 | 0.03 |
ENST00000595621.1
ENST00000595051.1 |
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr2_+_33661382 | 0.03 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr11_-_31832862 | 0.03 |
ENST00000379115.4
|
PAX6
|
paired box 6 |
| chr3_-_176914191 | 0.03 |
ENST00000437738.1
ENST00000424913.1 ENST00000443315.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr15_+_64386261 | 0.03 |
ENST00000560829.1
|
SNX1
|
sorting nexin 1 |
| chr16_+_67261008 | 0.03 |
ENST00000304800.9
ENST00000563953.1 ENST00000565201.1 |
TMEM208
|
transmembrane protein 208 |
| chr8_-_25281747 | 0.03 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr16_-_2205352 | 0.03 |
ENST00000563192.1
|
RP11-304L19.5
|
RP11-304L19.5 |
| chr4_+_86396321 | 0.03 |
ENST00000503995.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr16_+_31085714 | 0.03 |
ENST00000300850.5
ENST00000564189.1 ENST00000428260.1 |
ZNF646
|
zinc finger protein 646 |
| chr4_+_186064395 | 0.03 |
ENST00000281456.6
|
SLC25A4
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 |
| chr11_-_62389621 | 0.03 |
ENST00000531383.1
ENST00000265471.5 |
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr21_+_33671264 | 0.03 |
ENST00000339944.4
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr19_-_8567478 | 0.03 |
ENST00000255612.3
|
PRAM1
|
PML-RARA regulated adaptor molecule 1 |
| chr19_+_10959043 | 0.03 |
ENST00000397820.4
|
C19orf38
|
chromosome 19 open reading frame 38 |
| chr19_+_42387228 | 0.03 |
ENST00000354532.3
ENST00000599846.1 ENST00000347545.4 |
ARHGEF1
|
Rho guanine nucleotide exchange factor (GEF) 1 |
| chr2_+_109204909 | 0.03 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr3_-_114790179 | 0.03 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr3_-_46249878 | 0.03 |
ENST00000296140.3
|
CCR1
|
chemokine (C-C motif) receptor 1 |
| chr5_-_55412774 | 0.03 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr19_+_35739782 | 0.03 |
ENST00000347609.4
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr21_-_27107283 | 0.03 |
ENST00000284971.3
ENST00000400099.1 |
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr8_+_27491572 | 0.03 |
ENST00000301904.3
|
SCARA3
|
scavenger receptor class A, member 3 |
| chr14_+_63671105 | 0.03 |
ENST00000316754.3
|
RHOJ
|
ras homolog family member J |
| chr16_+_28874860 | 0.03 |
ENST00000545570.1
|
SH2B1
|
SH2B adaptor protein 1 |
| chr22_+_38004473 | 0.03 |
ENST00000414350.3
ENST00000343632.4 |
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr12_-_56652111 | 0.03 |
ENST00000267116.7
|
ANKRD52
|
ankyrin repeat domain 52 |
| chr19_+_35739897 | 0.03 |
ENST00000605618.1
ENST00000427250.1 ENST00000601623.1 |
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr2_-_163100045 | 0.03 |
ENST00000188790.4
|
FAP
|
fibroblast activation protein, alpha |
| chr22_+_38004832 | 0.03 |
ENST00000405147.3
ENST00000429218.1 ENST00000325180.8 ENST00000337437.4 |
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr12_-_53601055 | 0.03 |
ENST00000552972.1
ENST00000422257.3 ENST00000267082.5 |
ITGB7
|
integrin, beta 7 |
| chr16_-_66968265 | 0.03 |
ENST00000567511.1
ENST00000422424.2 |
FAM96B
|
family with sequence similarity 96, member B |
| chr12_-_120638902 | 0.03 |
ENST00000551150.1
ENST00000313104.5 ENST00000547191.1 ENST00000546989.1 ENST00000392514.4 ENST00000228306.4 ENST00000550856.1 |
RPLP0
|
ribosomal protein, large, P0 |
| chr19_-_4559814 | 0.03 |
ENST00000586582.1
|
SEMA6B
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
| chr6_+_32811885 | 0.03 |
ENST00000458296.1
ENST00000413039.1 ENST00000429600.1 ENST00000412095.1 ENST00000415067.1 ENST00000395330.1 |
TAPSAR1
PSMB9
|
TAP1 and PSMB8 antisense RNA 1 proteasome (prosome, macropain) subunit, beta type, 9 |
| chr2_-_175499294 | 0.03 |
ENST00000392547.2
|
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr17_-_38545799 | 0.03 |
ENST00000577541.1
|
TOP2A
|
topoisomerase (DNA) II alpha 170kDa |
| chr17_-_19651598 | 0.03 |
ENST00000570414.1
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr17_-_79269067 | 0.03 |
ENST00000288439.5
ENST00000374759.3 |
SLC38A10
|
solute carrier family 38, member 10 |
| chr3_-_129513259 | 0.03 |
ENST00000329333.5
|
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr21_+_17909594 | 0.03 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr12_-_123187890 | 0.03 |
ENST00000328880.5
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chr7_+_134551583 | 0.03 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr15_+_90544532 | 0.03 |
ENST00000268154.4
|
ZNF710
|
zinc finger protein 710 |
| chr6_-_6007200 | 0.03 |
ENST00000244766.2
|
NRN1
|
neuritin 1 |
| chr17_+_7462103 | 0.03 |
ENST00000396545.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr6_-_154677866 | 0.03 |
ENST00000367220.4
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr6_-_33267101 | 0.03 |
ENST00000497454.1
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr22_+_38004723 | 0.03 |
ENST00000381756.5
|
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr17_-_47022140 | 0.03 |
ENST00000290330.3
|
SNF8
|
SNF8, ESCRT-II complex subunit |
| chr5_+_61874696 | 0.03 |
ENST00000491184.2
|
LRRC70
|
leucine rich repeat containing 70 |
| chr16_-_28506840 | 0.03 |
ENST00000569430.1
|
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr16_-_21663950 | 0.03 |
ENST00000268389.4
|
IGSF6
|
immunoglobulin superfamily, member 6 |
| chr3_-_71114066 | 0.03 |
ENST00000485326.2
|
FOXP1
|
forkhead box P1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SPI1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.3 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.0 | GO:0014839 | myoblast migration involved in skeletal muscle regeneration(GO:0014839) |
| 0.0 | 0.1 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.0 | 0.1 | GO:0042704 | detection of oxygen(GO:0003032) uterine wall breakdown(GO:0042704) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.0 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.0 | GO:1902563 | regulation of neutrophil degranulation(GO:0043313) regulation of neutrophil activation(GO:1902563) |
| 0.0 | 0.0 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.0 | 0.0 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.0 | 0.1 | GO:0043375 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.0 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.1 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.1 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.0 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.0 | 0.1 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.0 | 0.1 | GO:0033087 | negative regulation of immature T cell proliferation(GO:0033087) |
| 0.0 | 0.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.0 | GO:0072737 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.0 | 0.1 | GO:0010716 | negative regulation of extracellular matrix disassembly(GO:0010716) |
| 0.0 | 0.1 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.0 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.0 | GO:0032242 | regulation of nucleoside transport(GO:0032242) |
| 0.0 | 0.1 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.0 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.2 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.0 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.1 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.0 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.0 | 0.0 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.2 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.0 | GO:0034681 | integrin alpha11-beta1 complex(GO:0034681) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.0 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.0 | 0.1 | GO:0034038 | deoxyhypusine synthase activity(GO:0034038) |
| 0.0 | 0.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.0 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.0 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.0 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |