Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
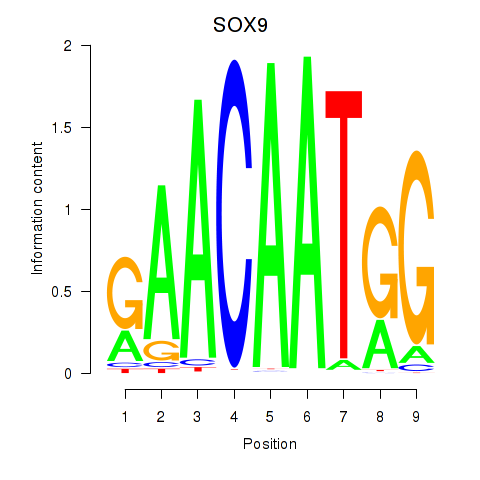
Results for SOX9
Z-value: 0.96
Transcription factors associated with SOX9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX9
|
ENSG00000125398.5 | SRY-box transcription factor 9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX9 | hg19_v2_chr17_+_70117153_70117174 | 0.37 | 6.3e-01 | Click! |
Activity profile of SOX9 motif
Sorted Z-values of SOX9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_61765732 | 0.65 |
ENST00000443240.1
ENST00000436018.1 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr1_-_156399184 | 0.54 |
ENST00000368243.1
ENST00000357975.4 ENST00000310027.5 ENST00000400991.2 |
C1orf61
|
chromosome 1 open reading frame 61 |
| chr12_+_95611536 | 0.54 |
ENST00000549002.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr2_-_240322685 | 0.52 |
ENST00000544989.1
|
HDAC4
|
histone deacetylase 4 |
| chr2_-_70475701 | 0.48 |
ENST00000282574.4
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr11_-_95522639 | 0.47 |
ENST00000536839.1
|
FAM76B
|
family with sequence similarity 76, member B |
| chr4_-_110723335 | 0.46 |
ENST00000394634.2
|
CFI
|
complement factor I |
| chr17_-_57232596 | 0.44 |
ENST00000581068.1
ENST00000330137.7 |
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr2_-_70475730 | 0.43 |
ENST00000445587.1
ENST00000433529.2 ENST00000415783.2 |
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chrX_+_78003204 | 0.42 |
ENST00000435339.3
ENST00000514744.1 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chr22_-_42739533 | 0.39 |
ENST00000515426.1
|
TCF20
|
transcription factor 20 (AR1) |
| chr7_+_129710350 | 0.38 |
ENST00000335420.5
ENST00000463413.1 |
KLHDC10
|
kelch domain containing 10 |
| chr10_-_99094458 | 0.38 |
ENST00000371019.2
|
FRAT2
|
frequently rearranged in advanced T-cell lymphomas 2 |
| chr1_-_93645818 | 0.38 |
ENST00000370280.1
ENST00000479918.1 |
TMED5
|
transmembrane emp24 protein transport domain containing 5 |
| chr4_+_95128748 | 0.37 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr1_+_173991633 | 0.37 |
ENST00000424181.1
|
RP11-160H22.3
|
RP11-160H22.3 |
| chr16_+_30934376 | 0.36 |
ENST00000562798.1
ENST00000471231.2 |
FBXL19
|
F-box and leucine-rich repeat protein 19 |
| chr5_+_40841410 | 0.35 |
ENST00000381677.3
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr1_+_228337553 | 0.34 |
ENST00000366714.2
|
GJC2
|
gap junction protein, gamma 2, 47kDa |
| chrX_-_106959631 | 0.34 |
ENST00000486554.1
ENST00000372390.4 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr6_-_86353510 | 0.34 |
ENST00000444272.1
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr2_+_46926326 | 0.34 |
ENST00000394861.2
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr2_-_175202151 | 0.33 |
ENST00000595354.1
|
AC018470.1
|
Uncharacterized protein FLJ46347 |
| chr2_-_160568918 | 0.33 |
ENST00000453016.1
ENST00000607836.1 |
AC009961.3
|
AC009961.3 |
| chr4_+_175204818 | 0.33 |
ENST00000503780.1
|
CEP44
|
centrosomal protein 44kDa |
| chr4_+_95128996 | 0.32 |
ENST00000457823.2
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr6_-_134861089 | 0.32 |
ENST00000606039.1
|
RP11-557H15.4
|
RP11-557H15.4 |
| chr6_-_27782548 | 0.32 |
ENST00000333151.3
|
HIST1H2AJ
|
histone cluster 1, H2aj |
| chr5_+_76012009 | 0.32 |
ENST00000505600.1
|
F2R
|
coagulation factor II (thrombin) receptor |
| chr1_-_243418650 | 0.31 |
ENST00000522995.1
|
CEP170
|
centrosomal protein 170kDa |
| chr15_+_98503922 | 0.31 |
ENST00000268042.6
|
ARRDC4
|
arrestin domain containing 4 |
| chr16_-_3068171 | 0.31 |
ENST00000572154.1
ENST00000328796.4 |
CLDN6
|
claudin 6 |
| chr2_-_37501692 | 0.31 |
ENST00000443977.1
|
PRKD3
|
protein kinase D3 |
| chr15_-_80695917 | 0.31 |
ENST00000559008.1
|
RP11-210M15.2
|
Uncharacterized protein |
| chr4_-_110723194 | 0.30 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr4_+_95129061 | 0.30 |
ENST00000354268.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr2_-_175869936 | 0.30 |
ENST00000409900.3
|
CHN1
|
chimerin 1 |
| chr20_+_48884002 | 0.30 |
ENST00000425497.1
ENST00000445003.1 |
RP11-290F20.3
|
RP11-290F20.3 |
| chr4_+_113152978 | 0.29 |
ENST00000309703.6
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr2_-_107503558 | 0.29 |
ENST00000361686.4
ENST00000409087.3 |
ST6GAL2
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2 |
| chr12_+_124457746 | 0.28 |
ENST00000392404.3
ENST00000538932.2 ENST00000337815.4 ENST00000540762.2 |
ZNF664
FAM101A
|
zinc finger protein 664 family with sequence similarity 101, member A |
| chr8_+_79578282 | 0.27 |
ENST00000263849.4
|
ZC2HC1A
|
zinc finger, C2HC-type containing 1A |
| chr2_-_61765315 | 0.27 |
ENST00000406957.1
ENST00000401558.2 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr14_+_102276192 | 0.26 |
ENST00000557714.1
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr11_-_111649015 | 0.26 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr18_+_7946839 | 0.25 |
ENST00000578916.1
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chr1_-_53018654 | 0.25 |
ENST00000257177.4
ENST00000355809.4 ENST00000528642.1 ENST00000470626.1 ENST00000371544.3 |
ZCCHC11
|
zinc finger, CCHC domain containing 11 |
| chr3_+_187871659 | 0.25 |
ENST00000416784.1
ENST00000430340.1 ENST00000414139.1 ENST00000454789.1 |
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr15_-_34502197 | 0.25 |
ENST00000557877.1
|
KATNBL1
|
katanin p80 subunit B-like 1 |
| chr6_+_64282447 | 0.25 |
ENST00000370650.2
ENST00000578299.1 |
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr3_+_181429704 | 0.25 |
ENST00000431565.2
ENST00000325404.1 |
SOX2
|
SRY (sex determining region Y)-box 2 |
| chr9_-_99064429 | 0.25 |
ENST00000375263.3
|
HSD17B3
|
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr9_-_130517522 | 0.25 |
ENST00000373274.3
ENST00000420366.1 |
SH2D3C
|
SH2 domain containing 3C |
| chr14_-_95236551 | 0.24 |
ENST00000238558.3
|
GSC
|
goosecoid homeobox |
| chr11_+_110225855 | 0.24 |
ENST00000526605.1
ENST00000526703.1 |
RP11-347E10.1
|
RP11-347E10.1 |
| chr1_-_221915418 | 0.23 |
ENST00000323825.3
ENST00000366899.3 |
DUSP10
|
dual specificity phosphatase 10 |
| chr1_-_26232951 | 0.23 |
ENST00000426559.2
ENST00000455785.2 |
STMN1
|
stathmin 1 |
| chr4_+_54243798 | 0.23 |
ENST00000337488.6
ENST00000358575.5 ENST00000507922.1 |
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr13_+_110958124 | 0.23 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr16_+_68573640 | 0.23 |
ENST00000398253.2
ENST00000573161.1 |
ZFP90
|
ZFP90 zinc finger protein |
| chr14_-_31495569 | 0.23 |
ENST00000357479.5
ENST00000355683.5 |
STRN3
|
striatin, calmodulin binding protein 3 |
| chr10_+_97803151 | 0.22 |
ENST00000403870.3
ENST00000265992.5 ENST00000465148.2 ENST00000534974.1 |
CCNJ
|
cyclin J |
| chr6_+_56911336 | 0.22 |
ENST00000370733.4
|
KIAA1586
|
KIAA1586 |
| chr6_+_100054606 | 0.22 |
ENST00000369215.4
|
PRDM13
|
PR domain containing 13 |
| chr10_+_91461337 | 0.22 |
ENST00000260753.4
ENST00000416354.1 ENST00000394289.2 ENST00000371728.3 |
KIF20B
|
kinesin family member 20B |
| chr18_-_24128496 | 0.22 |
ENST00000417602.1
|
KCTD1
|
potassium channel tetramerization domain containing 1 |
| chr4_+_54243862 | 0.22 |
ENST00000306932.6
|
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr6_+_89790459 | 0.21 |
ENST00000369472.1
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr2_-_158182322 | 0.21 |
ENST00000420719.2
ENST00000409216.1 |
ERMN
|
ermin, ERM-like protein |
| chr6_-_135375921 | 0.21 |
ENST00000367820.2
ENST00000314674.3 ENST00000524715.1 ENST00000415177.2 ENST00000367826.2 |
HBS1L
|
HBS1-like (S. cerevisiae) |
| chr10_+_69644404 | 0.21 |
ENST00000212015.6
|
SIRT1
|
sirtuin 1 |
| chr11_-_8892464 | 0.20 |
ENST00000527347.1
ENST00000526241.1 ENST00000526126.1 ENST00000530938.1 ENST00000526057.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr4_+_113152881 | 0.20 |
ENST00000274000.5
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr2_-_201828356 | 0.20 |
ENST00000234296.2
|
ORC2
|
origin recognition complex, subunit 2 |
| chr6_-_41040195 | 0.20 |
ENST00000463088.1
ENST00000469104.1 ENST00000486443.1 |
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr10_+_49514698 | 0.20 |
ENST00000432379.1
ENST00000429041.1 ENST00000374189.1 |
MAPK8
|
mitogen-activated protein kinase 8 |
| chrX_-_15872914 | 0.20 |
ENST00000380291.1
ENST00000545766.1 ENST00000421527.2 ENST00000329235.2 |
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr11_-_95522907 | 0.20 |
ENST00000358780.5
ENST00000542135.1 |
FAM76B
|
family with sequence similarity 76, member B |
| chr8_-_101965559 | 0.20 |
ENST00000353245.3
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr3_-_149688502 | 0.20 |
ENST00000481767.1
ENST00000475518.1 |
PFN2
|
profilin 2 |
| chr7_-_27239703 | 0.20 |
ENST00000222753.4
|
HOXA13
|
homeobox A13 |
| chr17_-_57232525 | 0.20 |
ENST00000583380.1
ENST00000580541.1 ENST00000578105.1 ENST00000437036.2 |
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr1_+_93913713 | 0.19 |
ENST00000604705.1
ENST00000370253.2 |
FNBP1L
|
formin binding protein 1-like |
| chr21_-_40720974 | 0.19 |
ENST00000380748.1
|
HMGN1
|
high mobility group nucleosome binding domain 1 |
| chr11_+_28724129 | 0.19 |
ENST00000513853.1
|
RP11-115J23.1
|
RP11-115J23.1 |
| chr3_-_182698381 | 0.19 |
ENST00000292782.4
|
DCUN1D1
|
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chr21_-_40720995 | 0.19 |
ENST00000380749.5
|
HMGN1
|
high mobility group nucleosome binding domain 1 |
| chr14_-_65569186 | 0.19 |
ENST00000555932.1
ENST00000358664.4 ENST00000284165.6 ENST00000358402.4 ENST00000246163.2 ENST00000556979.1 ENST00000555667.1 ENST00000557746.1 ENST00000556443.1 |
MAX
|
MYC associated factor X |
| chr1_+_24286287 | 0.19 |
ENST00000334351.7
ENST00000374468.1 |
PNRC2
|
proline-rich nuclear receptor coactivator 2 |
| chr7_-_11871815 | 0.19 |
ENST00000423059.4
|
THSD7A
|
thrombospondin, type I, domain containing 7A |
| chr2_+_74212073 | 0.19 |
ENST00000441217.1
|
AC073046.25
|
AC073046.25 |
| chr10_+_94050913 | 0.19 |
ENST00000358935.2
|
MARCH5
|
membrane-associated ring finger (C3HC4) 5 |
| chr10_+_70748487 | 0.19 |
ENST00000361983.4
|
KIAA1279
|
KIAA1279 |
| chr6_+_119215308 | 0.19 |
ENST00000229595.5
|
ASF1A
|
anti-silencing function 1A histone chaperone |
| chr19_+_7660716 | 0.19 |
ENST00000160298.4
ENST00000446248.2 |
CAMSAP3
|
calmodulin regulated spectrin-associated protein family, member 3 |
| chr17_-_16472483 | 0.19 |
ENST00000395824.1
ENST00000448349.2 ENST00000395825.3 |
ZNF287
|
zinc finger protein 287 |
| chr19_+_58514229 | 0.19 |
ENST00000546949.1
ENST00000553254.1 ENST00000547364.1 |
CTD-2368P22.1
|
HCG1811579; Uncharacterized protein |
| chr16_-_75590114 | 0.18 |
ENST00000568377.1
ENST00000565067.1 ENST00000258173.6 |
TMEM231
|
transmembrane protein 231 |
| chr18_-_44497308 | 0.18 |
ENST00000585916.1
ENST00000324794.7 ENST00000545673.1 |
PIAS2
|
protein inhibitor of activated STAT, 2 |
| chr7_+_77167376 | 0.18 |
ENST00000435495.2
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr6_-_24877490 | 0.17 |
ENST00000540914.1
ENST00000378023.4 |
FAM65B
|
family with sequence similarity 65, member B |
| chr18_-_74728998 | 0.17 |
ENST00000359645.3
ENST00000397875.3 ENST00000397869.3 ENST00000578193.1 ENST00000578873.1 ENST00000397866.4 ENST00000528160.1 ENST00000527041.1 ENST00000526111.1 ENST00000397865.5 ENST00000382582.3 |
MBP
|
myelin basic protein |
| chr12_-_88974236 | 0.17 |
ENST00000228280.5
ENST00000552044.1 ENST00000357116.4 |
KITLG
|
KIT ligand |
| chr7_+_94537542 | 0.17 |
ENST00000433881.1
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr7_-_107968999 | 0.17 |
ENST00000456431.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr12_-_122985067 | 0.17 |
ENST00000540586.1
ENST00000543897.1 |
ZCCHC8
|
zinc finger, CCHC domain containing 8 |
| chr14_-_103989033 | 0.17 |
ENST00000553878.1
ENST00000557530.1 |
CKB
|
creatine kinase, brain |
| chr1_+_244998918 | 0.17 |
ENST00000366528.3
|
COX20
|
COX20 cytochrome C oxidase assembly factor |
| chr5_+_172068232 | 0.17 |
ENST00000520919.1
ENST00000522853.1 ENST00000369800.5 |
NEURL1B
|
neuralized E3 ubiquitin protein ligase 1B |
| chr7_+_16793160 | 0.17 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr6_-_41040268 | 0.16 |
ENST00000373154.2
ENST00000244558.9 ENST00000464633.1 ENST00000424266.2 ENST00000479950.1 ENST00000482515.1 |
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr12_-_2966193 | 0.16 |
ENST00000382678.3
|
AC005841.1
|
Uncharacterized protein ENSP00000372125 |
| chr11_+_34073195 | 0.16 |
ENST00000341394.4
|
CAPRIN1
|
cell cycle associated protein 1 |
| chr22_+_38864041 | 0.16 |
ENST00000216014.4
ENST00000409006.3 |
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr1_-_243418621 | 0.16 |
ENST00000366544.1
ENST00000366543.1 |
CEP170
|
centrosomal protein 170kDa |
| chr6_-_64029879 | 0.16 |
ENST00000370658.5
ENST00000485906.2 ENST00000370657.4 |
LGSN
|
lengsin, lens protein with glutamine synthetase domain |
| chr11_-_102323740 | 0.16 |
ENST00000398136.2
|
TMEM123
|
transmembrane protein 123 |
| chr9_+_19408999 | 0.16 |
ENST00000340967.2
|
ACER2
|
alkaline ceramidase 2 |
| chr4_+_24661479 | 0.16 |
ENST00000569621.1
|
RP11-496D24.2
|
RP11-496D24.2 |
| chr4_-_110723134 | 0.16 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr9_-_123476612 | 0.16 |
ENST00000426959.1
|
MEGF9
|
multiple EGF-like-domains 9 |
| chrX_+_129473859 | 0.16 |
ENST00000424447.1
|
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr13_+_53029564 | 0.16 |
ENST00000468284.1
ENST00000378034.3 ENST00000258607.5 ENST00000378037.5 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr2_-_55277692 | 0.16 |
ENST00000394611.2
|
RTN4
|
reticulon 4 |
| chr1_+_185014496 | 0.16 |
ENST00000367510.3
|
RNF2
|
ring finger protein 2 |
| chr15_-_64673630 | 0.16 |
ENST00000558008.1
ENST00000559519.1 ENST00000380258.2 |
KIAA0101
|
KIAA0101 |
| chr4_+_2819883 | 0.15 |
ENST00000511747.1
ENST00000503393.2 |
SH3BP2
|
SH3-domain binding protein 2 |
| chr8_+_42195972 | 0.15 |
ENST00000532157.1
ENST00000265421.4 ENST00000520008.1 |
POLB
|
polymerase (DNA directed), beta |
| chr1_+_200708671 | 0.15 |
ENST00000358823.2
|
CAMSAP2
|
calmodulin regulated spectrin-associated protein family, member 2 |
| chr3_+_25469724 | 0.15 |
ENST00000437042.2
|
RARB
|
retinoic acid receptor, beta |
| chr13_-_107214291 | 0.15 |
ENST00000375926.1
|
ARGLU1
|
arginine and glutamate rich 1 |
| chr3_-_123339343 | 0.15 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr21_+_30671690 | 0.15 |
ENST00000399921.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr4_-_40516560 | 0.15 |
ENST00000513473.1
|
RBM47
|
RNA binding motif protein 47 |
| chr18_+_56530794 | 0.15 |
ENST00000590285.1
ENST00000586085.1 ENST00000589288.1 |
ZNF532
|
zinc finger protein 532 |
| chr3_-_128294929 | 0.15 |
ENST00000356020.2
|
C3orf27
|
chromosome 3 open reading frame 27 |
| chr16_-_15736881 | 0.15 |
ENST00000540441.2
|
KIAA0430
|
KIAA0430 |
| chr12_-_71003568 | 0.15 |
ENST00000547715.1
ENST00000451516.2 ENST00000538708.1 ENST00000550857.1 ENST00000261266.5 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chrX_+_84498989 | 0.15 |
ENST00000395402.1
|
ZNF711
|
zinc finger protein 711 |
| chr7_+_102389434 | 0.15 |
ENST00000409231.3
ENST00000418198.1 |
FAM185A
|
family with sequence similarity 185, member A |
| chr11_-_102323489 | 0.14 |
ENST00000361236.3
|
TMEM123
|
transmembrane protein 123 |
| chr16_-_53737722 | 0.14 |
ENST00000569716.1
ENST00000562588.1 ENST00000562230.1 ENST00000379925.3 ENST00000563746.1 ENST00000568653.3 |
RPGRIP1L
|
RPGRIP1-like |
| chr1_-_243418344 | 0.14 |
ENST00000366542.1
|
CEP170
|
centrosomal protein 170kDa |
| chr17_+_74723031 | 0.14 |
ENST00000586200.1
|
METTL23
|
methyltransferase like 23 |
| chr19_+_42254885 | 0.14 |
ENST00000595740.1
|
CEACAM6
|
carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) |
| chr11_+_125703204 | 0.14 |
ENST00000534411.1
ENST00000457514.2 |
PATE4
|
prostate and testis expressed 4 |
| chr3_-_149688896 | 0.14 |
ENST00000239940.7
|
PFN2
|
profilin 2 |
| chr20_-_1317555 | 0.14 |
ENST00000537552.1
|
AL136531.1
|
HCG2043693; Uncharacterized protein |
| chr3_+_25469802 | 0.14 |
ENST00000330688.4
|
RARB
|
retinoic acid receptor, beta |
| chr8_+_42196000 | 0.14 |
ENST00000518925.1
ENST00000538005.1 |
POLB
|
polymerase (DNA directed), beta |
| chr5_+_140593509 | 0.14 |
ENST00000341948.4
|
PCDHB13
|
protocadherin beta 13 |
| chr2_+_30569506 | 0.14 |
ENST00000421976.2
|
AC109642.1
|
AC109642.1 |
| chr2_-_55276320 | 0.14 |
ENST00000357376.3
|
RTN4
|
reticulon 4 |
| chr15_+_101402041 | 0.14 |
ENST00000558475.1
ENST00000558641.1 ENST00000559673.1 |
RP11-66B24.1
|
RP11-66B24.1 |
| chrX_-_135962876 | 0.14 |
ENST00000431446.3
ENST00000570135.1 ENST00000320676.7 ENST00000562646.1 |
RBMX
|
RNA binding motif protein, X-linked |
| chrX_+_84499038 | 0.14 |
ENST00000373165.3
|
ZNF711
|
zinc finger protein 711 |
| chr14_+_32546274 | 0.14 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr1_-_151689259 | 0.14 |
ENST00000420342.1
ENST00000290583.4 |
CELF3
|
CUGBP, Elav-like family member 3 |
| chr5_+_138677515 | 0.14 |
ENST00000265192.4
ENST00000511706.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr5_+_94890840 | 0.13 |
ENST00000504763.1
|
ARSK
|
arylsulfatase family, member K |
| chr14_-_69619823 | 0.13 |
ENST00000341516.5
|
DCAF5
|
DDB1 and CUL4 associated factor 5 |
| chr3_-_8686479 | 0.13 |
ENST00000544814.1
ENST00000427408.1 |
SSUH2
|
ssu-2 homolog (C. elegans) |
| chr1_+_14075865 | 0.13 |
ENST00000413440.1
|
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr4_-_76598326 | 0.13 |
ENST00000503660.1
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr1_-_184723942 | 0.13 |
ENST00000318130.8
|
EDEM3
|
ER degradation enhancer, mannosidase alpha-like 3 |
| chr14_+_102276209 | 0.13 |
ENST00000445439.3
ENST00000334743.5 ENST00000557095.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr6_-_127840048 | 0.13 |
ENST00000467753.1
|
SOGA3
|
SOGA family member 3 |
| chr9_-_85882145 | 0.13 |
ENST00000328788.1
|
FRMD3
|
FERM domain containing 3 |
| chr17_-_49124230 | 0.13 |
ENST00000510283.1
ENST00000510855.1 |
SPAG9
|
sperm associated antigen 9 |
| chr9_-_139658965 | 0.13 |
ENST00000316144.5
|
LCN15
|
lipocalin 15 |
| chr6_-_53213780 | 0.13 |
ENST00000304434.6
ENST00000370918.4 |
ELOVL5
|
ELOVL fatty acid elongase 5 |
| chr19_-_44123734 | 0.13 |
ENST00000598676.1
|
ZNF428
|
zinc finger protein 428 |
| chr2_+_232573222 | 0.13 |
ENST00000341369.7
ENST00000409683.1 |
PTMA
|
prothymosin, alpha |
| chr7_-_99277610 | 0.13 |
ENST00000343703.5
ENST00000222982.4 ENST00000439761.1 ENST00000339843.2 |
CYP3A5
|
cytochrome P450, family 3, subfamily A, polypeptide 5 |
| chr8_-_101965146 | 0.13 |
ENST00000395957.2
ENST00000395948.2 ENST00000457309.1 |
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr1_-_26232522 | 0.13 |
ENST00000399728.1
|
STMN1
|
stathmin 1 |
| chr9_-_99064386 | 0.13 |
ENST00000375262.2
|
HSD17B3
|
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr7_+_29519486 | 0.13 |
ENST00000409041.4
|
CHN2
|
chimerin 2 |
| chrX_+_107068959 | 0.13 |
ENST00000451923.1
|
MID2
|
midline 2 |
| chr4_-_2264015 | 0.13 |
ENST00000337190.2
|
MXD4
|
MAX dimerization protein 4 |
| chr17_+_59489112 | 0.13 |
ENST00000335108.2
|
C17orf82
|
chromosome 17 open reading frame 82 |
| chr15_+_89164560 | 0.13 |
ENST00000379231.3
ENST00000559528.1 |
AEN
|
apoptosis enhancing nuclease |
| chr19_+_45971246 | 0.12 |
ENST00000585836.1
ENST00000417353.2 ENST00000353609.3 ENST00000591858.1 ENST00000443841.2 ENST00000590335.1 |
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr12_-_122985494 | 0.12 |
ENST00000336229.4
|
ZCCHC8
|
zinc finger, CCHC domain containing 8 |
| chr14_+_93357749 | 0.12 |
ENST00000557689.1
|
RP11-862G15.1
|
RP11-862G15.1 |
| chr2_+_168725458 | 0.12 |
ENST00000392690.3
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr1_-_68698197 | 0.12 |
ENST00000370973.2
ENST00000370971.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr14_+_32546145 | 0.12 |
ENST00000556611.1
ENST00000539826.2 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr2_+_232573208 | 0.12 |
ENST00000409115.3
|
PTMA
|
prothymosin, alpha |
| chr10_-_15130767 | 0.12 |
ENST00000356189.5
|
ACBD7
|
acyl-CoA binding domain containing 7 |
| chr5_+_140557371 | 0.12 |
ENST00000239444.2
|
PCDHB8
|
protocadherin beta 8 |
| chr13_+_51913819 | 0.12 |
ENST00000419898.2
|
SERPINE3
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 3 |
| chr3_+_196466710 | 0.12 |
ENST00000327134.3
|
PAK2
|
p21 protein (Cdc42/Rac)-activated kinase 2 |
| chr9_+_80912059 | 0.12 |
ENST00000347159.2
ENST00000376588.3 |
PSAT1
|
phosphoserine aminotransferase 1 |
| chr6_-_27440837 | 0.12 |
ENST00000211936.6
|
ZNF184
|
zinc finger protein 184 |
| chr5_+_40679584 | 0.12 |
ENST00000302472.3
|
PTGER4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr7_+_69064300 | 0.12 |
ENST00000342771.4
|
AUTS2
|
autism susceptibility candidate 2 |
| chr4_-_83351294 | 0.12 |
ENST00000502762.1
|
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr17_+_7761301 | 0.12 |
ENST00000332439.4
ENST00000570446.1 |
CYB5D1
|
cytochrome b5 domain containing 1 |
| chr9_-_123476719 | 0.12 |
ENST00000373930.3
|
MEGF9
|
multiple EGF-like-domains 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX9
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.1 | 0.3 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.1 | 0.9 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.5 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 0.2 | GO:1990619 | histone H3-K9 deacetylation(GO:1990619) |
| 0.1 | 0.2 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.1 | 0.2 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.1 | 0.2 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.1 | 1.4 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 0.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.5 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.5 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.2 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.2 | GO:1900133 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) regulation of renin secretion into blood stream(GO:1900133) |
| 0.0 | 0.6 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.4 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.0 | 0.2 | GO:1904048 | positive regulation of long term synaptic depression(GO:1900454) regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.0 | 0.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.2 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.1 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 0.0 | 0.2 | GO:1903490 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.0 | 0.1 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.1 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.3 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.2 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.3 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.0 | 0.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.2 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.1 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.2 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.0 | 0.4 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.2 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.2 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.1 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.2 | GO:0001763 | morphogenesis of a branching structure(GO:0001763) |
| 0.0 | 0.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.2 | GO:0055096 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.0 | 0.2 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.3 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.0 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.3 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 0.4 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:1904617 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.0 | 0.1 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.2 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.4 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.1 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.1 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.0 | 0.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.2 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0097466 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) glycoprotein ERAD pathway(GO:0097466) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.0 | 0.2 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0001701 | in utero embryonic development(GO:0001701) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.3 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.2 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 | 0.1 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.1 | GO:0019374 | galactosylceramide metabolic process(GO:0006681) galactolipid metabolic process(GO:0019374) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.0 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.0 | 0.1 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.4 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.5 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.1 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.4 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.0 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.2 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.9 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) proline transmembrane transport(GO:0035524) |
| 0.0 | 0.2 | GO:0043584 | nose development(GO:0043584) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 0.2 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.1 | 0.9 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.2 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.3 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.6 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.2 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.1 | 0.2 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.1 | 0.2 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.1 | 0.2 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.2 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.1 | 0.5 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.1 | 0.4 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.1 | 0.9 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.2 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.5 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.3 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 0.3 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.2 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.4 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.9 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.1 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.4 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.0 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.1 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.2 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.2 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.3 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.0 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.1 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.3 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.9 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.9 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |