Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
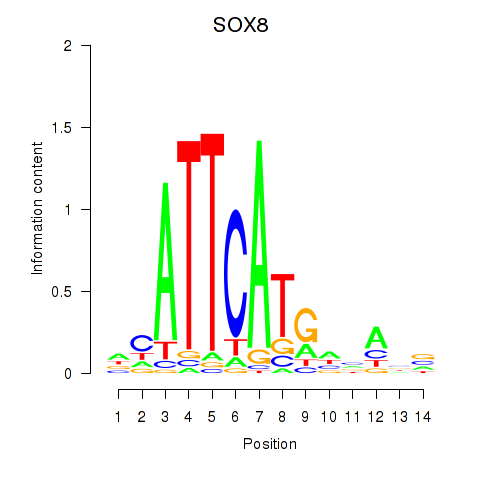
Results for SOX8
Z-value: 0.38
Transcription factors associated with SOX8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX8
|
ENSG00000005513.9 | SRY-box transcription factor 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX8 | hg19_v2_chr16_+_1031762_1031808 | 0.25 | 7.5e-01 | Click! |
Activity profile of SOX8 motif
Sorted Z-values of SOX8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_52076425 | 0.42 |
ENST00000436511.2
|
ZNF175
|
zinc finger protein 175 |
| chr21_-_47352477 | 0.42 |
ENST00000593412.1
|
PRED62
|
Uncharacterized protein |
| chr20_-_5426380 | 0.24 |
ENST00000609252.1
ENST00000422352.1 |
LINC00658
|
long intergenic non-protein coding RNA 658 |
| chr2_-_187367356 | 0.23 |
ENST00000595956.1
|
AC018867.2
|
AC018867.2 |
| chr6_+_37012607 | 0.17 |
ENST00000423336.1
|
COX6A1P2
|
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
| chr12_+_113354341 | 0.14 |
ENST00000553152.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr2_+_233527443 | 0.14 |
ENST00000410095.1
|
EFHD1
|
EF-hand domain family, member D1 |
| chr12_-_10007448 | 0.13 |
ENST00000538152.1
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr4_-_75695366 | 0.13 |
ENST00000512743.1
|
BTC
|
betacellulin |
| chr22_+_24236191 | 0.13 |
ENST00000215754.7
|
MIF
|
macrophage migration inhibitory factor (glycosylation-inhibiting factor) |
| chr9_-_95244781 | 0.12 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr7_+_100303676 | 0.11 |
ENST00000303151.4
|
POP7
|
processing of precursor 7, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr17_-_7145475 | 0.11 |
ENST00000571129.1
ENST00000571253.1 ENST00000573928.1 |
GABARAP
|
GABA(A) receptor-associated protein |
| chr14_+_52327350 | 0.11 |
ENST00000555472.1
ENST00000556766.1 |
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr1_+_153330322 | 0.11 |
ENST00000368738.3
|
S100A9
|
S100 calcium binding protein A9 |
| chr20_+_32250079 | 0.11 |
ENST00000375222.3
|
C20orf144
|
chromosome 20 open reading frame 144 |
| chr20_+_48807351 | 0.11 |
ENST00000303004.3
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta |
| chr4_-_47983519 | 0.11 |
ENST00000358519.4
ENST00000544810.1 ENST00000402813.3 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr4_-_75024085 | 0.10 |
ENST00000600169.1
|
AC093677.1
|
Uncharacterized protein |
| chr6_-_31689456 | 0.10 |
ENST00000495859.1
ENST00000375819.2 |
LY6G6C
|
lymphocyte antigen 6 complex, locus G6C |
| chr17_-_34207295 | 0.09 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr11_+_18154059 | 0.09 |
ENST00000531264.1
|
MRGPRX3
|
MAS-related GPR, member X3 |
| chr2_-_25451065 | 0.09 |
ENST00000606328.1
|
RP11-458N5.1
|
RP11-458N5.1 |
| chr1_+_8378140 | 0.09 |
ENST00000377479.2
|
SLC45A1
|
solute carrier family 45, member 1 |
| chr4_-_140544386 | 0.08 |
ENST00000561977.1
|
RP11-308D13.3
|
RP11-308D13.3 |
| chr3_-_49851313 | 0.08 |
ENST00000333486.3
|
UBA7
|
ubiquitin-like modifier activating enzyme 7 |
| chr10_-_128110441 | 0.08 |
ENST00000456514.1
|
LINC00601
|
long intergenic non-protein coding RNA 601 |
| chr12_+_14927270 | 0.08 |
ENST00000544848.1
|
H2AFJ
|
H2A histone family, member J |
| chr6_+_80129989 | 0.07 |
ENST00000429444.1
|
RP1-232L24.3
|
RP1-232L24.3 |
| chr18_-_51750948 | 0.07 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr22_-_20255212 | 0.07 |
ENST00000416372.1
|
RTN4R
|
reticulon 4 receptor |
| chr12_-_123187890 | 0.07 |
ENST00000328880.5
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chr2_+_166326157 | 0.07 |
ENST00000421875.1
ENST00000314499.7 ENST00000409664.1 |
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr3_+_11267691 | 0.07 |
ENST00000413416.1
|
HRH1
|
histamine receptor H1 |
| chr4_+_75174204 | 0.07 |
ENST00000332112.4
ENST00000514968.1 ENST00000503098.1 ENST00000502358.1 ENST00000509145.1 ENST00000505212.1 |
EPGN
|
epithelial mitogen |
| chr11_-_58035732 | 0.07 |
ENST00000395079.2
|
OR10W1
|
olfactory receptor, family 10, subfamily W, member 1 |
| chr19_+_36606933 | 0.06 |
ENST00000586868.1
|
TBCB
|
tubulin folding cofactor B |
| chr2_+_7118755 | 0.06 |
ENST00000433456.1
|
RNF144A
|
ring finger protein 144A |
| chr1_-_200992827 | 0.06 |
ENST00000332129.2
ENST00000422435.2 |
KIF21B
|
kinesin family member 21B |
| chr1_+_161475208 | 0.06 |
ENST00000367972.4
ENST00000271450.6 |
FCGR2A
|
Fc fragment of IgG, low affinity IIa, receptor (CD32) |
| chr2_+_39117010 | 0.06 |
ENST00000409978.1
|
ARHGEF33
|
Rho guanine nucleotide exchange factor (GEF) 33 |
| chr6_-_49712147 | 0.06 |
ENST00000433368.2
ENST00000354620.4 |
CRISP3
|
cysteine-rich secretory protein 3 |
| chr7_-_47578840 | 0.06 |
ENST00000450444.1
|
TNS3
|
tensin 3 |
| chr2_-_239140011 | 0.06 |
ENST00000409376.1
ENST00000409070.1 ENST00000409942.1 |
AC016757.3
|
Protein LOC151174 |
| chr1_+_26496362 | 0.06 |
ENST00000374266.5
ENST00000270812.5 |
ZNF593
|
zinc finger protein 593 |
| chr2_-_151395525 | 0.06 |
ENST00000439275.1
|
RND3
|
Rho family GTPase 3 |
| chr9_-_16276311 | 0.06 |
ENST00000380685.1
|
C9orf92
|
chromosome 9 open reading frame 92 |
| chr21_-_19858196 | 0.06 |
ENST00000422787.1
|
TMPRSS15
|
transmembrane protease, serine 15 |
| chr10_-_131762105 | 0.06 |
ENST00000368648.3
ENST00000355311.5 |
EBF3
|
early B-cell factor 3 |
| chr2_-_197675000 | 0.06 |
ENST00000342506.2
|
C2orf66
|
chromosome 2 open reading frame 66 |
| chr11_-_125366089 | 0.06 |
ENST00000366139.3
ENST00000278919.3 |
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr3_+_157154578 | 0.06 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr17_+_68047418 | 0.06 |
ENST00000586373.1
ENST00000588782.1 |
LINC01028
|
long intergenic non-protein coding RNA 1028 |
| chr2_-_120124258 | 0.06 |
ENST00000409877.1
ENST00000409523.1 ENST00000409466.2 ENST00000414534.1 |
C2orf76
|
chromosome 2 open reading frame 76 |
| chr10_-_375422 | 0.06 |
ENST00000434695.2
|
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
| chr19_+_39903185 | 0.06 |
ENST00000409794.3
|
PLEKHG2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr10_+_88414298 | 0.05 |
ENST00000372071.2
|
OPN4
|
opsin 4 |
| chr17_+_22022437 | 0.05 |
ENST00000540040.1
|
MTRNR2L1
|
MT-RNR2-like 1 |
| chr2_-_70944855 | 0.05 |
ENST00000415348.1
|
ADD2
|
adducin 2 (beta) |
| chr3_-_146213722 | 0.05 |
ENST00000336685.2
ENST00000489015.1 |
PLSCR2
|
phospholipid scramblase 2 |
| chr4_+_75174180 | 0.05 |
ENST00000413830.1
|
EPGN
|
epithelial mitogen |
| chr12_+_1099675 | 0.05 |
ENST00000545318.2
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr19_+_1524072 | 0.05 |
ENST00000454744.2
|
PLK5
|
polo-like kinase 5 |
| chr5_-_160973649 | 0.05 |
ENST00000393959.1
ENST00000517547.1 |
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr7_-_122342988 | 0.05 |
ENST00000434824.1
|
RNF148
|
ring finger protein 148 |
| chr2_-_228497888 | 0.05 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chr15_+_62853562 | 0.05 |
ENST00000561311.1
|
TLN2
|
talin 2 |
| chr10_+_103986085 | 0.05 |
ENST00000370005.3
|
ELOVL3
|
ELOVL fatty acid elongase 3 |
| chrX_-_53461305 | 0.05 |
ENST00000168216.6
|
HSD17B10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr11_+_57435219 | 0.05 |
ENST00000527985.1
ENST00000287169.3 |
ZDHHC5
|
zinc finger, DHHC-type containing 5 |
| chr19_+_51728316 | 0.05 |
ENST00000436584.2
ENST00000421133.2 ENST00000391796.3 ENST00000262262.4 |
CD33
|
CD33 molecule |
| chr4_+_89299994 | 0.05 |
ENST00000264346.7
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr14_-_101053739 | 0.05 |
ENST00000554140.1
|
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr2_+_173792893 | 0.05 |
ENST00000535187.1
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr9_+_44868935 | 0.05 |
ENST00000448436.2
|
RP11-160N1.10
|
RP11-160N1.10 |
| chrX_-_53461288 | 0.05 |
ENST00000375298.4
ENST00000375304.5 |
HSD17B10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr5_-_94417314 | 0.05 |
ENST00000505208.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr2_+_7073174 | 0.05 |
ENST00000416587.1
|
RNF144A
|
ring finger protein 144A |
| chr4_-_137020391 | 0.05 |
ENST00000569694.1
|
RP11-775H9.3
|
RP11-775H9.3 |
| chr3_+_173116225 | 0.05 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr5_+_156887027 | 0.05 |
ENST00000435489.2
ENST00000311946.7 |
NIPAL4
|
NIPA-like domain containing 4 |
| chr21_-_43735628 | 0.05 |
ENST00000291525.10
ENST00000518498.1 |
TFF3
|
trefoil factor 3 (intestinal) |
| chr17_-_8021710 | 0.05 |
ENST00000380149.1
ENST00000448843.2 |
ALOXE3
|
arachidonate lipoxygenase 3 |
| chr3_+_191046810 | 0.05 |
ENST00000392455.3
ENST00000392456.3 |
CCDC50
|
coiled-coil domain containing 50 |
| chr19_-_42806919 | 0.05 |
ENST00000595530.1
ENST00000538771.1 ENST00000601865.1 |
PAFAH1B3
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr19_+_45681997 | 0.05 |
ENST00000433642.2
|
BLOC1S3
|
biogenesis of lysosomal organelles complex-1, subunit 3 |
| chrX_+_48687283 | 0.05 |
ENST00000338270.1
|
ERAS
|
ES cell expressed Ras |
| chr13_+_108921977 | 0.05 |
ENST00000430559.1
ENST00000375887.4 |
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr6_-_150217195 | 0.05 |
ENST00000532335.1
|
RAET1E
|
retinoic acid early transcript 1E |
| chr14_+_35591928 | 0.04 |
ENST00000605870.1
ENST00000557404.3 |
KIAA0391
|
KIAA0391 |
| chr6_+_42018614 | 0.04 |
ENST00000465926.1
ENST00000482432.1 |
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa |
| chr5_+_140602904 | 0.04 |
ENST00000515856.2
ENST00000239449.4 |
PCDHB14
|
protocadherin beta 14 |
| chr15_+_86087267 | 0.04 |
ENST00000558166.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr12_+_133066137 | 0.04 |
ENST00000434748.2
|
FBRSL1
|
fibrosin-like 1 |
| chr1_+_110577229 | 0.04 |
ENST00000369795.3
ENST00000369794.2 |
STRIP1
|
striatin interacting protein 1 |
| chr4_-_10686475 | 0.04 |
ENST00000226951.6
|
CLNK
|
cytokine-dependent hematopoietic cell linker |
| chr21_-_16125773 | 0.04 |
ENST00000454128.2
|
AF127936.3
|
AF127936.3 |
| chr14_+_88471468 | 0.04 |
ENST00000267549.3
|
GPR65
|
G protein-coupled receptor 65 |
| chr12_+_29376673 | 0.04 |
ENST00000547116.1
|
FAR2
|
fatty acyl CoA reductase 2 |
| chr10_-_4285923 | 0.04 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr5_+_140227048 | 0.04 |
ENST00000532602.1
|
PCDHA9
|
protocadherin alpha 9 |
| chr3_-_176914191 | 0.04 |
ENST00000437738.1
ENST00000424913.1 ENST00000443315.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr1_-_238649319 | 0.04 |
ENST00000400946.2
|
RP11-371I1.2
|
long intergenic non-protein coding RNA 1139 |
| chr4_-_113627966 | 0.04 |
ENST00000505632.1
|
RP11-148B6.2
|
RP11-148B6.2 |
| chr3_-_193272741 | 0.04 |
ENST00000392443.3
|
ATP13A4
|
ATPase type 13A4 |
| chr8_-_82359662 | 0.04 |
ENST00000519260.1
ENST00000256103.2 |
PMP2
|
peripheral myelin protein 2 |
| chr12_+_122242597 | 0.04 |
ENST00000267197.5
|
SETD1B
|
SET domain containing 1B |
| chr2_+_97203082 | 0.04 |
ENST00000454558.2
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr1_-_222763240 | 0.04 |
ENST00000352967.4
ENST00000391882.1 ENST00000543857.1 |
TAF1A
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa |
| chr11_-_33743952 | 0.04 |
ENST00000534312.1
|
CD59
|
CD59 molecule, complement regulatory protein |
| chr1_+_200993071 | 0.04 |
ENST00000446333.1
ENST00000458003.1 |
RP11-168O16.1
|
RP11-168O16.1 |
| chr16_-_28503357 | 0.04 |
ENST00000333496.9
ENST00000561505.1 ENST00000567963.1 ENST00000354630.5 ENST00000355477.5 ENST00000357076.5 ENST00000565688.1 ENST00000359984.7 |
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr7_+_80275752 | 0.04 |
ENST00000419819.2
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr13_+_98794810 | 0.04 |
ENST00000595437.1
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr2_-_202645612 | 0.04 |
ENST00000409632.2
ENST00000410052.1 ENST00000467448.1 |
ALS2
|
amyotrophic lateral sclerosis 2 (juvenile) |
| chr15_+_45926919 | 0.04 |
ENST00000561735.1
ENST00000260324.7 |
SQRDL
|
sulfide quinone reductase-like (yeast) |
| chr1_+_112016414 | 0.04 |
ENST00000343534.5
ENST00000369718.3 |
C1orf162
|
chromosome 1 open reading frame 162 |
| chr3_-_54962100 | 0.04 |
ENST00000273286.5
|
LRTM1
|
leucine-rich repeats and transmembrane domains 1 |
| chr2_-_37501692 | 0.04 |
ENST00000443977.1
|
PRKD3
|
protein kinase D3 |
| chr12_+_29376592 | 0.04 |
ENST00000182377.4
|
FAR2
|
fatty acyl CoA reductase 2 |
| chr12_-_50101003 | 0.04 |
ENST00000550488.1
|
FMNL3
|
formin-like 3 |
| chr17_+_34087888 | 0.04 |
ENST00000586491.1
ENST00000588628.1 ENST00000285023.4 |
C17orf50
|
chromosome 17 open reading frame 50 |
| chr22_-_30722912 | 0.04 |
ENST00000215790.7
|
TBC1D10A
|
TBC1 domain family, member 10A |
| chr22_+_45714301 | 0.04 |
ENST00000427777.1
|
FAM118A
|
family with sequence similarity 118, member A |
| chr18_-_3845293 | 0.04 |
ENST00000400145.2
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr19_+_22235279 | 0.04 |
ENST00000594363.1
ENST00000597927.1 ENST00000594947.1 |
ZNF257
|
zinc finger protein 257 |
| chr22_-_30642728 | 0.03 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr2_-_158485387 | 0.03 |
ENST00000243349.8
|
ACVR1C
|
activin A receptor, type IC |
| chr6_-_32160622 | 0.03 |
ENST00000487761.1
ENST00000375040.3 |
GPSM3
|
G-protein signaling modulator 3 |
| chr15_+_25101698 | 0.03 |
ENST00000400097.1
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N |
| chr1_+_182584314 | 0.03 |
ENST00000566297.1
|
RP11-317P15.4
|
RP11-317P15.4 |
| chr8_+_1993152 | 0.03 |
ENST00000262113.4
|
MYOM2
|
myomesin 2 |
| chr18_-_29264669 | 0.03 |
ENST00000306851.5
|
B4GALT6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr6_-_51952367 | 0.03 |
ENST00000340994.4
|
PKHD1
|
polycystic kidney and hepatic disease 1 (autosomal recessive) |
| chr17_-_41132410 | 0.03 |
ENST00000409446.3
ENST00000453594.1 ENST00000409399.1 ENST00000421990.2 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr17_-_37123646 | 0.03 |
ENST00000378079.2
|
FBXO47
|
F-box protein 47 |
| chr19_+_18794470 | 0.03 |
ENST00000321949.8
ENST00000338797.6 |
CRTC1
|
CREB regulated transcription coactivator 1 |
| chr5_-_94417186 | 0.03 |
ENST00000312216.8
ENST00000512425.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr3_-_169587621 | 0.03 |
ENST00000523069.1
ENST00000316428.5 ENST00000264676.5 |
LRRC31
|
leucine rich repeat containing 31 |
| chr22_-_50523760 | 0.03 |
ENST00000395876.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr3_+_173302222 | 0.03 |
ENST00000361589.4
|
NLGN1
|
neuroligin 1 |
| chr3_+_38347427 | 0.03 |
ENST00000273173.4
|
SLC22A14
|
solute carrier family 22, member 14 |
| chr12_-_112856623 | 0.03 |
ENST00000551291.2
|
RPL6
|
ribosomal protein L6 |
| chr11_-_62995986 | 0.03 |
ENST00000403374.2
|
SLC22A25
|
solute carrier family 22, member 25 |
| chr6_-_52668605 | 0.03 |
ENST00000334575.5
|
GSTA1
|
glutathione S-transferase alpha 1 |
| chr16_+_87985029 | 0.03 |
ENST00000439677.1
ENST00000286122.7 ENST00000355163.5 ENST00000454563.1 ENST00000479780.2 ENST00000393208.2 ENST00000412691.1 ENST00000355022.4 |
BANP
|
BTG3 associated nuclear protein |
| chr10_-_21186144 | 0.03 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr10_+_53806501 | 0.03 |
ENST00000373975.2
|
PRKG1
|
protein kinase, cGMP-dependent, type I |
| chr16_+_82068873 | 0.03 |
ENST00000566213.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr3_-_21792838 | 0.03 |
ENST00000281523.2
|
ZNF385D
|
zinc finger protein 385D |
| chr3_+_577893 | 0.03 |
ENST00000420823.1
ENST00000442809.1 |
AC090044.1
|
AC090044.1 |
| chr9_+_99690592 | 0.03 |
ENST00000354649.3
|
NUTM2G
|
NUT family member 2G |
| chr20_+_42543504 | 0.03 |
ENST00000341197.4
|
TOX2
|
TOX high mobility group box family member 2 |
| chr12_-_12714025 | 0.03 |
ENST00000539940.1
|
DUSP16
|
dual specificity phosphatase 16 |
| chr12_-_9760482 | 0.03 |
ENST00000229402.3
|
KLRB1
|
killer cell lectin-like receptor subfamily B, member 1 |
| chr2_+_204732487 | 0.03 |
ENST00000302823.3
|
CTLA4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr20_+_32399093 | 0.03 |
ENST00000217402.2
|
CHMP4B
|
charged multivesicular body protein 4B |
| chr15_-_58306295 | 0.03 |
ENST00000559517.1
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr16_+_14280742 | 0.03 |
ENST00000341243.5
|
MKL2
|
MKL/myocardin-like 2 |
| chr22_-_30722866 | 0.03 |
ENST00000403477.3
|
TBC1D10A
|
TBC1 domain family, member 10A |
| chr6_+_116832789 | 0.03 |
ENST00000368599.3
|
FAM26E
|
family with sequence similarity 26, member E |
| chr3_-_141719195 | 0.03 |
ENST00000397991.4
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr13_-_49987885 | 0.03 |
ENST00000409082.1
|
CAB39L
|
calcium binding protein 39-like |
| chr19_-_37663332 | 0.03 |
ENST00000392157.2
|
ZNF585A
|
zinc finger protein 585A |
| chr12_-_123201337 | 0.03 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr5_+_140792614 | 0.03 |
ENST00000398610.2
|
PCDHGA10
|
protocadherin gamma subfamily A, 10 |
| chr16_-_122619 | 0.03 |
ENST00000262316.6
|
RHBDF1
|
rhomboid 5 homolog 1 (Drosophila) |
| chr17_+_7387677 | 0.03 |
ENST00000322644.6
|
POLR2A
|
polymerase (RNA) II (DNA directed) polypeptide A, 220kDa |
| chr11_+_122733011 | 0.03 |
ENST00000533709.1
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr9_-_130661916 | 0.03 |
ENST00000373142.1
ENST00000373146.1 ENST00000373144.3 |
ST6GALNAC6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr1_-_85930246 | 0.03 |
ENST00000426972.3
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr19_-_37697976 | 0.03 |
ENST00000588873.1
|
CTC-454I21.3
|
Uncharacterized protein; Zinc finger protein 585B |
| chr8_+_1993173 | 0.03 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr8_+_125283924 | 0.03 |
ENST00000517482.1
|
RP11-383J24.1
|
RP11-383J24.1 |
| chr10_-_116418053 | 0.03 |
ENST00000277895.5
|
ABLIM1
|
actin binding LIM protein 1 |
| chr5_-_138534071 | 0.03 |
ENST00000394817.2
|
SIL1
|
SIL1 nucleotide exchange factor |
| chr19_+_1269324 | 0.03 |
ENST00000589710.1
ENST00000588230.1 ENST00000413636.2 ENST00000586472.1 ENST00000589686.1 ENST00000444172.2 ENST00000587323.1 ENST00000320936.5 ENST00000587896.1 ENST00000589235.1 ENST00000591659.1 |
CIRBP
|
cold inducible RNA binding protein |
| chr7_+_18548878 | 0.03 |
ENST00000456174.2
|
HDAC9
|
histone deacetylase 9 |
| chr1_-_9811600 | 0.03 |
ENST00000435891.1
|
CLSTN1
|
calsyntenin 1 |
| chr11_-_121986923 | 0.03 |
ENST00000560104.1
|
BLID
|
BH3-like motif containing, cell death inducer |
| chr4_-_16085657 | 0.03 |
ENST00000543373.1
|
PROM1
|
prominin 1 |
| chr3_+_35683651 | 0.03 |
ENST00000187397.4
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr9_-_114557207 | 0.03 |
ENST00000374287.3
ENST00000374283.5 |
C9orf84
|
chromosome 9 open reading frame 84 |
| chr1_-_152297679 | 0.03 |
ENST00000368799.1
|
FLG
|
filaggrin |
| chr17_-_66951474 | 0.03 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr3_+_130150307 | 0.03 |
ENST00000512836.1
|
COL6A5
|
collagen, type VI, alpha 5 |
| chr11_+_67219867 | 0.03 |
ENST00000438189.2
|
CABP4
|
calcium binding protein 4 |
| chr11_+_12308447 | 0.03 |
ENST00000256186.2
|
MICALCL
|
MICAL C-terminal like |
| chrX_-_50386648 | 0.03 |
ENST00000460112.3
|
SHROOM4
|
shroom family member 4 |
| chr4_+_159131596 | 0.03 |
ENST00000512481.1
|
TMEM144
|
transmembrane protein 144 |
| chr12_+_79371565 | 0.03 |
ENST00000551304.1
|
SYT1
|
synaptotagmin I |
| chr12_+_78359999 | 0.03 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr1_-_109935819 | 0.02 |
ENST00000538502.1
|
SORT1
|
sortilin 1 |
| chr15_+_78830023 | 0.02 |
ENST00000599596.1
|
AC027228.1
|
HCG2005369; Uncharacterized protein |
| chr1_-_183538319 | 0.02 |
ENST00000420553.1
ENST00000419402.1 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr17_-_2169425 | 0.02 |
ENST00000570606.1
ENST00000354901.4 |
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr1_+_153631130 | 0.02 |
ENST00000368685.5
|
SNAPIN
|
SNAP-associated protein |
| chr4_+_74347400 | 0.02 |
ENST00000226355.3
|
AFM
|
afamin |
| chr18_+_71815743 | 0.02 |
ENST00000169551.6
ENST00000580087.1 |
TIMM21
|
translocase of inner mitochondrial membrane 21 homolog (yeast) |
| chr4_+_169633310 | 0.02 |
ENST00000510998.1
|
PALLD
|
palladin, cytoskeletal associated protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX8
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.1 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.1 | GO:0002906 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.1 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.1 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.0 | 0.1 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.0 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.0 | GO:0006186 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.0 | 0.1 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0000035 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.1 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.0 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.0 | 0.0 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.1 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |