Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
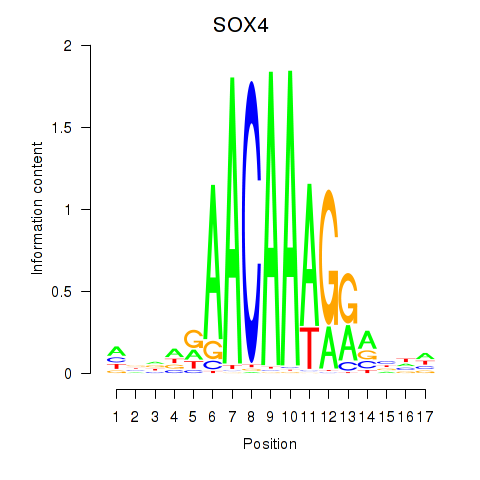
Results for SOX4
Z-value: 0.59
Transcription factors associated with SOX4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX4
|
ENSG00000124766.4 | SRY-box transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX4 | hg19_v2_chr6_+_21593972_21594071 | 0.37 | 6.3e-01 | Click! |
Activity profile of SOX4 motif
Sorted Z-values of SOX4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_18948208 | 0.66 |
ENST00000607346.1
|
ARL5B-AS1
|
ARL5B antisense RNA 1 |
| chr11_-_10829851 | 0.37 |
ENST00000532082.1
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr4_-_170192000 | 0.36 |
ENST00000502315.1
|
SH3RF1
|
SH3 domain containing ring finger 1 |
| chr15_+_91416092 | 0.35 |
ENST00000559353.1
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chr1_+_207002222 | 0.34 |
ENST00000270218.6
|
IL19
|
interleukin 19 |
| chr2_-_1748214 | 0.34 |
ENST00000433670.1
ENST00000425171.1 ENST00000252804.4 |
PXDN
|
peroxidasin homolog (Drosophila) |
| chr7_-_27219849 | 0.31 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr11_-_10830463 | 0.30 |
ENST00000527419.1
ENST00000530211.1 ENST00000530702.1 ENST00000524932.1 ENST00000532570.1 |
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr16_-_48281305 | 0.29 |
ENST00000356608.2
|
ABCC11
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 11 |
| chr22_-_31688431 | 0.28 |
ENST00000402249.3
ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr15_-_76304731 | 0.25 |
ENST00000394907.3
|
NRG4
|
neuregulin 4 |
| chr7_-_35293740 | 0.25 |
ENST00000408931.3
|
TBX20
|
T-box 20 |
| chr13_+_98795664 | 0.25 |
ENST00000376581.5
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr1_-_155880672 | 0.23 |
ENST00000609492.1
ENST00000368322.3 |
RIT1
|
Ras-like without CAAX 1 |
| chrX_-_73512411 | 0.23 |
ENST00000602576.1
ENST00000429124.1 |
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chr17_+_61473104 | 0.23 |
ENST00000583016.1
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr14_-_21516590 | 0.23 |
ENST00000555026.1
|
NDRG2
|
NDRG family member 2 |
| chr4_-_75024085 | 0.22 |
ENST00000600169.1
|
AC093677.1
|
Uncharacterized protein |
| chr2_+_208423891 | 0.22 |
ENST00000448277.1
ENST00000457101.1 |
CREB1
|
cAMP responsive element binding protein 1 |
| chr15_-_74495188 | 0.22 |
ENST00000563965.1
ENST00000395105.4 |
STRA6
|
stimulated by retinoic acid 6 |
| chr2_-_114647327 | 0.22 |
ENST00000602760.1
|
RP11-141B14.1
|
RP11-141B14.1 |
| chr11_+_5711010 | 0.22 |
ENST00000454828.1
|
TRIM22
|
tripartite motif containing 22 |
| chr7_+_99699280 | 0.21 |
ENST00000421755.1
|
AP4M1
|
adaptor-related protein complex 4, mu 1 subunit |
| chr22_+_37959647 | 0.21 |
ENST00000415670.1
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chr9_+_125486269 | 0.20 |
ENST00000259466.1
|
OR1L4
|
olfactory receptor, family 1, subfamily L, member 4 |
| chr15_+_63335899 | 0.20 |
ENST00000561266.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr22_-_31688381 | 0.20 |
ENST00000487265.2
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr22_+_31477296 | 0.20 |
ENST00000426927.1
ENST00000440425.1 ENST00000358743.1 ENST00000347557.2 ENST00000333137.7 |
SMTN
|
smoothelin |
| chr19_-_38720294 | 0.19 |
ENST00000412732.1
ENST00000456296.1 |
DPF1
|
D4, zinc and double PHD fingers family 1 |
| chr17_+_64961026 | 0.19 |
ENST00000262138.3
|
CACNG4
|
calcium channel, voltage-dependent, gamma subunit 4 |
| chr10_+_6244829 | 0.19 |
ENST00000317350.4
ENST00000379785.1 ENST00000379782.3 ENST00000360521.2 ENST00000379775.4 |
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr7_-_105926058 | 0.19 |
ENST00000417537.1
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr17_+_16945820 | 0.19 |
ENST00000577514.1
|
MPRIP
|
myosin phosphatase Rho interacting protein |
| chr8_-_42358742 | 0.19 |
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr15_-_41166414 | 0.18 |
ENST00000220507.4
|
RHOV
|
ras homolog family member V |
| chr5_+_102201430 | 0.18 |
ENST00000438793.3
ENST00000346918.2 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr16_+_30406721 | 0.18 |
ENST00000320159.2
|
ZNF48
|
zinc finger protein 48 |
| chrX_+_12993202 | 0.18 |
ENST00000451311.2
ENST00000380636.1 |
TMSB4X
|
thymosin beta 4, X-linked |
| chr1_+_212797789 | 0.17 |
ENST00000294829.3
|
FAM71A
|
family with sequence similarity 71, member A |
| chr2_+_220309379 | 0.17 |
ENST00000451076.1
|
SPEG
|
SPEG complex locus |
| chr14_-_61124977 | 0.17 |
ENST00000554986.1
|
SIX1
|
SIX homeobox 1 |
| chr8_+_9911778 | 0.17 |
ENST00000317173.4
ENST00000441698.2 |
MSRA
|
methionine sulfoxide reductase A |
| chr3_-_171489085 | 0.17 |
ENST00000418087.1
|
PLD1
|
phospholipase D1, phosphatidylcholine-specific |
| chr5_+_140480083 | 0.16 |
ENST00000231130.2
|
PCDHB3
|
protocadherin beta 3 |
| chr6_-_133055815 | 0.16 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr7_+_99699179 | 0.16 |
ENST00000438383.1
ENST00000429084.1 ENST00000359593.4 ENST00000439416.1 |
AP4M1
|
adaptor-related protein complex 4, mu 1 subunit |
| chr3_-_134092561 | 0.16 |
ENST00000510560.1
ENST00000504234.1 ENST00000515172.1 |
AMOTL2
|
angiomotin like 2 |
| chr17_+_80416050 | 0.16 |
ENST00000579198.1
ENST00000390006.4 ENST00000580296.1 |
NARF
|
nuclear prelamin A recognition factor |
| chr2_+_201981663 | 0.16 |
ENST00000433445.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr11_-_71810258 | 0.16 |
ENST00000544594.1
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr13_+_111855399 | 0.15 |
ENST00000426768.2
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr7_-_127671674 | 0.15 |
ENST00000478726.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr8_+_9911945 | 0.15 |
ENST00000518255.1
|
MSRA
|
methionine sulfoxide reductase A |
| chr22_-_36357671 | 0.15 |
ENST00000408983.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr8_+_61822605 | 0.15 |
ENST00000526936.1
|
AC022182.1
|
AC022182.1 |
| chr12_+_93963590 | 0.14 |
ENST00000340600.2
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr20_-_56284816 | 0.14 |
ENST00000395819.3
ENST00000341744.3 |
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr12_+_27623565 | 0.14 |
ENST00000535986.1
|
SMCO2
|
single-pass membrane protein with coiled-coil domains 2 |
| chr7_-_148581251 | 0.14 |
ENST00000478654.1
ENST00000460911.1 ENST00000350995.2 |
EZH2
|
enhancer of zeste homolog 2 (Drosophila) |
| chr12_-_10282836 | 0.14 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr6_+_123038689 | 0.14 |
ENST00000354275.2
ENST00000368446.1 |
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr21_+_30672433 | 0.13 |
ENST00000451655.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chrX_+_12993336 | 0.13 |
ENST00000380635.1
|
TMSB4X
|
thymosin beta 4, X-linked |
| chr5_+_102201509 | 0.13 |
ENST00000348126.2
ENST00000379787.4 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr1_+_153940713 | 0.13 |
ENST00000368601.1
ENST00000368603.1 ENST00000368600.3 |
CREB3L4
|
cAMP responsive element binding protein 3-like 4 |
| chr7_-_99699538 | 0.12 |
ENST00000343023.6
ENST00000303887.5 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr14_+_69658480 | 0.12 |
ENST00000409949.1
ENST00000409242.1 ENST00000312994.5 ENST00000413191.1 |
EXD2
|
exonuclease 3'-5' domain containing 2 |
| chr17_+_26698677 | 0.12 |
ENST00000457710.3
|
SARM1
|
sterile alpha and TIR motif containing 1 |
| chr1_+_153940346 | 0.12 |
ENST00000405694.3
ENST00000449724.1 ENST00000368607.3 ENST00000271889.4 |
CREB3L4
|
cAMP responsive element binding protein 3-like 4 |
| chr14_+_35761580 | 0.12 |
ENST00000553809.1
ENST00000555764.1 ENST00000556506.1 |
PSMA6
|
proteasome (prosome, macropain) subunit, alpha type, 6 |
| chr7_+_150076406 | 0.12 |
ENST00000329630.5
|
ZNF775
|
zinc finger protein 775 |
| chr17_+_41561317 | 0.12 |
ENST00000540306.1
ENST00000262415.3 ENST00000605777.1 |
DHX8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr10_+_11206925 | 0.12 |
ENST00000354440.2
ENST00000315874.4 ENST00000427450.1 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr3_+_141596371 | 0.12 |
ENST00000495216.1
|
ATP1B3
|
ATPase, Na+/K+ transporting, beta 3 polypeptide |
| chr7_-_127672146 | 0.12 |
ENST00000476782.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr13_-_24007815 | 0.12 |
ENST00000382298.3
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr15_+_25200108 | 0.12 |
ENST00000577949.1
ENST00000338094.6 ENST00000338327.4 ENST00000579070.1 ENST00000577565.1 |
SNURF
SNRPN
|
SNRPN upstream reading frame protein small nuclear ribonucleoprotein polypeptide N |
| chr4_-_54424366 | 0.12 |
ENST00000306888.2
|
LNX1
|
ligand of numb-protein X 1, E3 ubiquitin protein ligase |
| chr1_+_47137445 | 0.12 |
ENST00000569393.1
ENST00000334122.4 ENST00000415500.1 |
TEX38
|
testis expressed 38 |
| chr4_+_118955500 | 0.12 |
ENST00000296499.5
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr16_-_68482440 | 0.11 |
ENST00000219334.5
|
SMPD3
|
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II) |
| chr15_-_70390191 | 0.11 |
ENST00000559191.1
|
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr20_+_60698180 | 0.11 |
ENST00000361670.3
|
LSM14B
|
LSM14B, SCD6 homolog B (S. cerevisiae) |
| chr5_-_65018834 | 0.11 |
ENST00000506816.1
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr20_+_45338126 | 0.11 |
ENST00000359271.2
|
SLC2A10
|
solute carrier family 2 (facilitated glucose transporter), member 10 |
| chr4_-_22444733 | 0.11 |
ENST00000508133.1
|
GPR125
|
G protein-coupled receptor 125 |
| chr4_-_69111401 | 0.11 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr14_-_71276211 | 0.11 |
ENST00000381250.4
ENST00000555993.2 |
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9 |
| chr5_-_173043591 | 0.11 |
ENST00000285908.5
ENST00000480951.1 ENST00000311086.4 |
BOD1
|
biorientation of chromosomes in cell division 1 |
| chr5_-_146833222 | 0.11 |
ENST00000534907.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr5_+_150816595 | 0.10 |
ENST00000520111.1
ENST00000520701.1 ENST00000429484.2 |
SLC36A1
|
solute carrier family 36 (proton/amino acid symporter), member 1 |
| chr1_-_153940097 | 0.10 |
ENST00000413622.1
ENST00000310483.6 |
SLC39A1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr9_+_112852477 | 0.10 |
ENST00000480388.1
|
AKAP2
|
A kinase (PRKA) anchor protein 2 |
| chr11_-_85430356 | 0.10 |
ENST00000526999.1
|
SYTL2
|
synaptotagmin-like 2 |
| chr5_+_140235469 | 0.10 |
ENST00000506939.2
ENST00000307360.5 |
PCDHA10
|
protocadherin alpha 10 |
| chr19_-_38720354 | 0.10 |
ENST00000416611.1
|
DPF1
|
D4, zinc and double PHD fingers family 1 |
| chr17_+_67498396 | 0.10 |
ENST00000588110.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr16_-_48281444 | 0.10 |
ENST00000537808.1
ENST00000569991.1 |
ABCC11
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 11 |
| chr9_+_101705893 | 0.10 |
ENST00000375001.3
|
COL15A1
|
collagen, type XV, alpha 1 |
| chr22_-_42336209 | 0.10 |
ENST00000472374.2
|
CENPM
|
centromere protein M |
| chr19_-_55690758 | 0.10 |
ENST00000590851.1
|
SYT5
|
synaptotagmin V |
| chr17_+_80416482 | 0.10 |
ENST00000309794.11
ENST00000345415.7 ENST00000457415.3 ENST00000584411.1 ENST00000412079.2 ENST00000577432.1 |
NARF
|
nuclear prelamin A recognition factor |
| chr8_+_24151620 | 0.10 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr6_+_139456226 | 0.10 |
ENST00000367658.2
|
HECA
|
headcase homolog (Drosophila) |
| chr7_-_107643567 | 0.10 |
ENST00000393559.1
ENST00000439976.1 ENST00000393560.1 |
LAMB1
|
laminin, beta 1 |
| chr17_+_67498295 | 0.10 |
ENST00000589295.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr16_+_14980632 | 0.09 |
ENST00000565655.1
|
NOMO1
|
NODAL modulator 1 |
| chrX_-_153599578 | 0.09 |
ENST00000360319.4
ENST00000344736.4 |
FLNA
|
filamin A, alpha |
| chr12_-_109915098 | 0.09 |
ENST00000542858.1
ENST00000542262.1 ENST00000424763.2 |
KCTD10
|
potassium channel tetramerization domain containing 10 |
| chr9_-_117150243 | 0.09 |
ENST00000374088.3
|
AKNA
|
AT-hook transcription factor |
| chr15_+_75940218 | 0.09 |
ENST00000308527.5
|
SNX33
|
sorting nexin 33 |
| chr2_-_106015527 | 0.09 |
ENST00000344213.4
ENST00000358129.4 |
FHL2
|
four and a half LIM domains 2 |
| chr15_+_41136263 | 0.09 |
ENST00000568823.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chrX_-_73513307 | 0.09 |
ENST00000602420.1
|
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chr15_+_25200074 | 0.09 |
ENST00000390687.4
ENST00000584968.1 ENST00000346403.6 ENST00000554227.2 |
SNRPN
|
small nuclear ribonucleoprotein polypeptide N |
| chr3_-_38992052 | 0.09 |
ENST00000302328.3
ENST00000450244.1 ENST00000444237.2 |
SCN11A
|
sodium channel, voltage-gated, type XI, alpha subunit |
| chr5_+_140787600 | 0.09 |
ENST00000520790.1
|
PCDHGB6
|
protocadherin gamma subfamily B, 6 |
| chr6_-_132834184 | 0.09 |
ENST00000367941.2
ENST00000367937.4 |
STX7
|
syntaxin 7 |
| chr16_-_73082274 | 0.08 |
ENST00000268489.5
|
ZFHX3
|
zinc finger homeobox 3 |
| chr12_-_122296755 | 0.08 |
ENST00000289004.4
|
HPD
|
4-hydroxyphenylpyruvate dioxygenase |
| chr20_-_4804244 | 0.08 |
ENST00000379400.3
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2 |
| chr5_-_76788298 | 0.08 |
ENST00000511036.1
|
WDR41
|
WD repeat domain 41 |
| chrX_-_15683147 | 0.08 |
ENST00000380342.3
|
TMEM27
|
transmembrane protein 27 |
| chr14_+_101299520 | 0.08 |
ENST00000455531.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr7_-_148581360 | 0.08 |
ENST00000320356.2
ENST00000541220.1 ENST00000483967.1 ENST00000536783.1 |
EZH2
|
enhancer of zeste homolog 2 (Drosophila) |
| chr1_-_85930246 | 0.08 |
ENST00000426972.3
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr14_+_24439148 | 0.08 |
ENST00000543805.1
ENST00000534993.1 |
DHRS4L2
|
dehydrogenase/reductase (SDR family) member 4 like 2 |
| chr14_-_23446900 | 0.08 |
ENST00000556731.1
|
AJUBA
|
ajuba LIM protein |
| chrX_-_110655306 | 0.08 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chrX_+_51927919 | 0.08 |
ENST00000416960.1
|
MAGED4
|
melanoma antigen family D, 4 |
| chr2_-_208031542 | 0.08 |
ENST00000423015.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr3_-_128207349 | 0.08 |
ENST00000487848.1
|
GATA2
|
GATA binding protein 2 |
| chr16_-_58663720 | 0.08 |
ENST00000564557.1
ENST00000569240.1 ENST00000441024.2 ENST00000569020.1 ENST00000317147.5 |
CNOT1
|
CCR4-NOT transcription complex, subunit 1 |
| chr19_+_9296279 | 0.08 |
ENST00000344248.2
|
OR7D2
|
olfactory receptor, family 7, subfamily D, member 2 |
| chr20_+_18125727 | 0.07 |
ENST00000489634.2
|
CSRP2BP
|
CSRP2 binding protein |
| chr7_-_148725733 | 0.07 |
ENST00000286091.4
|
PDIA4
|
protein disulfide isomerase family A, member 4 |
| chr10_-_99393208 | 0.07 |
ENST00000307450.6
|
MORN4
|
MORN repeat containing 4 |
| chr2_-_127977654 | 0.07 |
ENST00000409327.1
|
CYP27C1
|
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr7_-_107643674 | 0.07 |
ENST00000222399.6
|
LAMB1
|
laminin, beta 1 |
| chr2_+_172543919 | 0.07 |
ENST00000452242.1
ENST00000340296.4 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr4_-_106629796 | 0.07 |
ENST00000416543.1
ENST00000515819.1 ENST00000420368.2 ENST00000503746.1 ENST00000340139.5 ENST00000433009.1 |
INTS12
|
integrator complex subunit 12 |
| chr12_-_49581152 | 0.07 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr1_-_161015752 | 0.07 |
ENST00000435396.1
ENST00000368021.3 |
USF1
|
upstream transcription factor 1 |
| chr6_-_139613269 | 0.07 |
ENST00000358430.3
|
TXLNB
|
taxilin beta |
| chr19_+_58095501 | 0.07 |
ENST00000536878.2
ENST00000597850.1 ENST00000597219.1 ENST00000598689.1 ENST00000599456.1 ENST00000307468.4 |
ZIK1
|
zinc finger protein interacting with K protein 1 |
| chr21_-_33104367 | 0.07 |
ENST00000286835.7
ENST00000399804.1 |
SCAF4
|
SR-related CTD-associated factor 4 |
| chr18_+_3450161 | 0.06 |
ENST00000551402.1
ENST00000577543.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr9_-_74383302 | 0.06 |
ENST00000377066.5
|
TMEM2
|
transmembrane protein 2 |
| chr15_+_81293254 | 0.06 |
ENST00000267984.2
|
MESDC1
|
mesoderm development candidate 1 |
| chr6_+_126240442 | 0.06 |
ENST00000448104.1
ENST00000438495.1 ENST00000444128.1 |
NCOA7
|
nuclear receptor coactivator 7 |
| chr4_-_186877502 | 0.06 |
ENST00000431902.1
ENST00000284776.7 ENST00000415274.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr4_-_184243561 | 0.06 |
ENST00000514470.1
ENST00000541814.1 |
CLDN24
|
claudin 24 |
| chr1_+_40840320 | 0.06 |
ENST00000372708.1
|
SMAP2
|
small ArfGAP2 |
| chr15_+_93426514 | 0.06 |
ENST00000556722.1
|
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr15_+_41136216 | 0.06 |
ENST00000562057.1
ENST00000344051.4 |
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr7_-_91794590 | 0.06 |
ENST00000430130.2
ENST00000454089.2 |
LRRD1
|
leucine-rich repeats and death domain containing 1 |
| chr12_-_99288536 | 0.06 |
ENST00000549797.1
ENST00000333732.7 ENST00000341752.7 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr10_+_22605304 | 0.06 |
ENST00000475460.2
ENST00000602390.1 ENST00000489125.2 ENST00000456711.1 ENST00000444869.1 |
COMMD3-BMI1
COMMD3
|
COMMD3-BMI1 readthrough COMM domain containing 3 |
| chr12_-_49582593 | 0.06 |
ENST00000295766.5
|
TUBA1A
|
tubulin, alpha 1a |
| chr14_+_91581011 | 0.06 |
ENST00000523894.1
ENST00000522322.1 ENST00000523771.1 |
C14orf159
|
chromosome 14 open reading frame 159 |
| chr3_+_57875711 | 0.06 |
ENST00000442599.2
|
SLMAP
|
sarcolemma associated protein |
| chr17_+_67498538 | 0.06 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr12_+_109915179 | 0.06 |
ENST00000434735.2
|
UBE3B
|
ubiquitin protein ligase E3B |
| chr2_-_136594740 | 0.06 |
ENST00000264162.2
|
LCT
|
lactase |
| chr11_-_85397167 | 0.06 |
ENST00000316398.3
|
CCDC89
|
coiled-coil domain containing 89 |
| chr14_+_91580708 | 0.06 |
ENST00000518868.1
|
C14orf159
|
chromosome 14 open reading frame 159 |
| chr1_+_44870866 | 0.06 |
ENST00000355387.2
ENST00000361799.2 |
RNF220
|
ring finger protein 220 |
| chr10_-_91102410 | 0.05 |
ENST00000282673.4
|
LIPA
|
lipase A, lysosomal acid, cholesterol esterase |
| chrX_-_110655391 | 0.05 |
ENST00000356915.2
ENST00000356220.3 |
DCX
|
doublecortin |
| chr12_-_49582837 | 0.05 |
ENST00000547939.1
ENST00000546918.1 ENST00000552924.1 |
TUBA1A
|
tubulin, alpha 1a |
| chr3_+_39509163 | 0.05 |
ENST00000436143.2
ENST00000441980.2 ENST00000311042.6 |
MOBP
|
myelin-associated oligodendrocyte basic protein |
| chr3_-_197024394 | 0.05 |
ENST00000434148.1
ENST00000412364.2 ENST00000436682.1 ENST00000456699.2 ENST00000392380.2 |
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr14_+_32798462 | 0.05 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr18_+_3449821 | 0.05 |
ENST00000407501.2
ENST00000405385.3 ENST00000546979.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr4_-_109089573 | 0.05 |
ENST00000265165.1
|
LEF1
|
lymphoid enhancer-binding factor 1 |
| chr7_-_42276612 | 0.05 |
ENST00000395925.3
ENST00000437480.1 |
GLI3
|
GLI family zinc finger 3 |
| chr10_-_61900762 | 0.05 |
ENST00000355288.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr1_+_47799542 | 0.05 |
ENST00000471289.2
ENST00000450808.2 |
CMPK1
|
cytidine monophosphate (UMP-CMP) kinase 1, cytosolic |
| chr3_+_39509070 | 0.05 |
ENST00000354668.4
ENST00000428261.1 ENST00000420739.1 ENST00000415443.1 ENST00000447324.1 ENST00000383754.3 |
MOBP
|
myelin-associated oligodendrocyte basic protein |
| chr9_-_131940526 | 0.05 |
ENST00000372491.2
|
IER5L
|
immediate early response 5-like |
| chr2_-_20425158 | 0.05 |
ENST00000381150.1
|
SDC1
|
syndecan 1 |
| chr6_+_22146851 | 0.05 |
ENST00000606197.1
|
CASC15
|
cancer susceptibility candidate 15 (non-protein coding) |
| chr12_+_95611516 | 0.05 |
ENST00000436874.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr22_-_50524298 | 0.05 |
ENST00000311597.5
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr14_-_73493825 | 0.04 |
ENST00000318876.5
ENST00000556143.1 |
ZFYVE1
|
zinc finger, FYVE domain containing 1 |
| chr12_-_10251603 | 0.04 |
ENST00000457018.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr13_-_110959478 | 0.04 |
ENST00000543140.1
ENST00000375820.4 |
COL4A1
|
collagen, type IV, alpha 1 |
| chr1_+_151682909 | 0.04 |
ENST00000326413.3
ENST00000442233.2 |
RIIAD1
AL589765.1
|
regulatory subunit of type II PKA R-subunit (RIIa) domain containing 1 Uncharacterized protein; cDNA FLJ36032 fis, clone TESTI2017069 |
| chr4_-_109090106 | 0.04 |
ENST00000379951.2
|
LEF1
|
lymphoid enhancer-binding factor 1 |
| chr2_+_54684327 | 0.04 |
ENST00000389980.3
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr2_-_75745823 | 0.04 |
ENST00000452003.1
|
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr18_+_7946941 | 0.04 |
ENST00000444013.1
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chr3_-_99833333 | 0.04 |
ENST00000354552.3
ENST00000331335.5 ENST00000398326.2 |
FILIP1L
|
filamin A interacting protein 1-like |
| chr12_-_95611149 | 0.04 |
ENST00000549499.1
ENST00000343958.4 ENST00000546711.1 |
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr14_+_24521202 | 0.04 |
ENST00000334420.7
ENST00000342740.5 |
LRRC16B
|
leucine rich repeat containing 16B |
| chr6_+_13925098 | 0.04 |
ENST00000488300.1
ENST00000544682.1 ENST00000420478.2 |
RNF182
|
ring finger protein 182 |
| chrX_+_101470280 | 0.04 |
ENST00000395088.2
ENST00000330252.5 ENST00000333110.5 |
NXF2
TCP11X1
|
nuclear RNA export factor 2 t-complex 11 family, X-linked 1 |
| chr22_-_21984282 | 0.04 |
ENST00000398873.3
ENST00000292778.6 |
YDJC
|
YdjC homolog (bacterial) |
| chr3_+_111260980 | 0.04 |
ENST00000438817.2
|
CD96
|
CD96 molecule |
| chr12_-_10251576 | 0.04 |
ENST00000315330.4
|
CLEC1A
|
C-type lectin domain family 1, member A |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0090472 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.1 | 0.5 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.2 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.1 | 0.2 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.2 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.1 | 0.3 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.3 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.2 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.0 | 0.3 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.2 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.0 | 0.2 | GO:2000909 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.1 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.1 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.2 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.1 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.0 | 0.0 | GO:0043585 | nose morphogenesis(GO:0043585) |
| 0.0 | 0.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.3 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.0 | 0.0 | GO:0048627 | myoblast development(GO:0048627) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.4 | GO:0015865 | purine nucleotide transport(GO:0015865) |
| 0.0 | 0.1 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.0 | 0.2 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.2 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.4 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.3 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0034128 | negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) |
| 0.0 | 0.1 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.0 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.0 | 0.0 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.2 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.2 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) proline transmembrane transport(GO:0035524) |
| 0.0 | 0.3 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.2 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043257 | laminin-8 complex(GO:0043257) |
| 0.0 | 0.4 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.0 | GO:0036457 | keratohyalin granule(GO:0036457) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.2 | GO:0032059 | muscle thin filament tropomyosin(GO:0005862) bleb(GO:0032059) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.2 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.1 | 0.3 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.3 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.4 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.3 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.2 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.4 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.2 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.0 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) L-proline transmembrane transporter activity(GO:0015193) alanine transmembrane transporter activity(GO:0022858) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |