Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
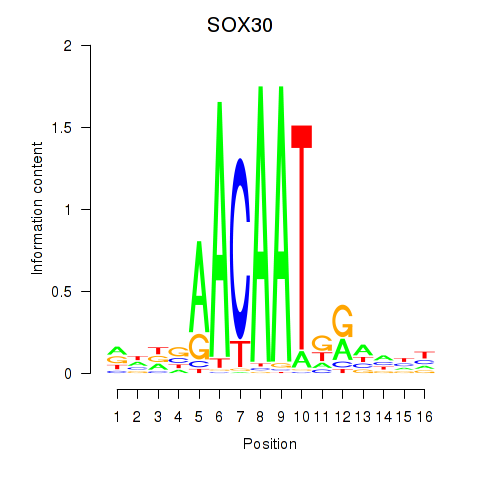
Results for SOX30
Z-value: 1.19
Transcription factors associated with SOX30
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX30
|
ENSG00000039600.6 | SRY-box transcription factor 30 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX30 | hg19_v2_chr5_-_157079372_157079395 | -0.60 | 4.0e-01 | Click! |
Activity profile of SOX30 motif
Sorted Z-values of SOX30 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_100770328 | 1.40 |
ENST00000223095.4
ENST00000445463.2 |
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr3_-_122283424 | 0.84 |
ENST00000477522.2
ENST00000360356.2 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr22_-_30970498 | 0.82 |
ENST00000431313.1
|
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr22_+_31488433 | 0.82 |
ENST00000455608.1
|
SMTN
|
smoothelin |
| chr5_-_146461027 | 0.79 |
ENST00000394410.2
ENST00000508267.1 ENST00000504198.1 |
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr14_+_101299520 | 0.64 |
ENST00000455531.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr4_+_54243798 | 0.61 |
ENST00000337488.6
ENST00000358575.5 ENST00000507922.1 |
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr2_-_158182322 | 0.60 |
ENST00000420719.2
ENST00000409216.1 |
ERMN
|
ermin, ERM-like protein |
| chr12_-_10588539 | 0.59 |
ENST00000381902.2
ENST00000381901.1 ENST00000539033.1 |
KLRC2
NKG2-E
|
killer cell lectin-like receptor subfamily C, member 2 Uncharacterized protein |
| chr22_-_30970560 | 0.59 |
ENST00000402369.1
ENST00000406361.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr1_+_149239529 | 0.55 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chr17_+_67498295 | 0.52 |
ENST00000589295.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr2_+_232573208 | 0.52 |
ENST00000409115.3
|
PTMA
|
prothymosin, alpha |
| chr6_-_134639180 | 0.51 |
ENST00000367858.5
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr1_-_201476274 | 0.50 |
ENST00000340006.2
|
CSRP1
|
cysteine and glycine-rich protein 1 |
| chr20_-_5426380 | 0.50 |
ENST00000609252.1
ENST00000422352.1 |
LINC00658
|
long intergenic non-protein coding RNA 658 |
| chr4_-_176733897 | 0.49 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr12_+_93096619 | 0.49 |
ENST00000397833.3
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr14_-_36645674 | 0.48 |
ENST00000556013.2
|
PTCSC3
|
papillary thyroid carcinoma susceptibility candidate 3 (non-protein coding) |
| chr12_+_9066472 | 0.47 |
ENST00000538657.1
|
PHC1
|
polyhomeotic homolog 1 (Drosophila) |
| chr12_+_93096759 | 0.46 |
ENST00000544406.2
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr9_-_134145880 | 0.46 |
ENST00000372269.3
ENST00000464831.1 |
FAM78A
|
family with sequence similarity 78, member A |
| chr11_+_46402744 | 0.46 |
ENST00000533952.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr22_+_46449674 | 0.43 |
ENST00000381051.2
|
FLJ27365
|
hsa-mir-4763 |
| chr2_+_143635067 | 0.40 |
ENST00000264170.4
|
KYNU
|
kynureninase |
| chr4_+_54243862 | 0.39 |
ENST00000306932.6
|
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr5_+_179135246 | 0.38 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr2_+_232573222 | 0.36 |
ENST00000341369.7
ENST00000409683.1 |
PTMA
|
prothymosin, alpha |
| chr18_+_68002675 | 0.35 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr12_+_27623565 | 0.34 |
ENST00000535986.1
|
SMCO2
|
single-pass membrane protein with coiled-coil domains 2 |
| chr15_+_76629064 | 0.34 |
ENST00000290759.4
|
ISL2
|
ISL LIM homeobox 2 |
| chr6_-_161085291 | 0.32 |
ENST00000316300.5
|
LPA
|
lipoprotein, Lp(a) |
| chr1_+_244998602 | 0.31 |
ENST00000411948.2
|
COX20
|
COX20 cytochrome C oxidase assembly factor |
| chr11_-_89224638 | 0.30 |
ENST00000535633.1
ENST00000263317.4 |
NOX4
|
NADPH oxidase 4 |
| chr1_-_21503337 | 0.30 |
ENST00000400422.1
ENST00000602326.1 ENST00000411888.1 ENST00000438975.1 |
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr7_+_870547 | 0.28 |
ENST00000457598.1
|
SUN1
|
Sad1 and UNC84 domain containing 1 |
| chr12_+_95611536 | 0.28 |
ENST00000549002.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr2_+_99953816 | 0.28 |
ENST00000289371.6
|
EIF5B
|
eukaryotic translation initiation factor 5B |
| chr3_+_19988736 | 0.27 |
ENST00000443878.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr2_+_179184955 | 0.27 |
ENST00000315022.2
|
OSBPL6
|
oxysterol binding protein-like 6 |
| chr1_+_244998918 | 0.26 |
ENST00000366528.3
|
COX20
|
COX20 cytochrome C oxidase assembly factor |
| chr10_+_35484793 | 0.26 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr12_+_11905413 | 0.25 |
ENST00000545027.1
|
ETV6
|
ets variant 6 |
| chr22_-_43485381 | 0.25 |
ENST00000331018.7
ENST00000266254.7 ENST00000445824.1 |
TTLL1
|
tubulin tyrosine ligase-like family, member 1 |
| chr4_+_113152978 | 0.25 |
ENST00000309703.6
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr6_+_131958436 | 0.24 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr14_+_52456193 | 0.23 |
ENST00000261700.3
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr6_-_49712123 | 0.23 |
ENST00000263045.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr5_+_140593509 | 0.23 |
ENST00000341948.4
|
PCDHB13
|
protocadherin beta 13 |
| chr12_-_99288986 | 0.22 |
ENST00000552407.1
ENST00000551613.1 ENST00000548447.1 ENST00000546364.3 ENST00000552748.1 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr1_+_180897269 | 0.22 |
ENST00000367587.1
|
KIAA1614
|
KIAA1614 |
| chr2_-_158182410 | 0.22 |
ENST00000419116.2
ENST00000410096.1 |
ERMN
|
ermin, ERM-like protein |
| chrX_+_102192200 | 0.20 |
ENST00000218249.5
|
RAB40AL
|
RAB40A, member RAS oncogene family-like |
| chr6_+_126240442 | 0.20 |
ENST00000448104.1
ENST00000438495.1 ENST00000444128.1 |
NCOA7
|
nuclear receptor coactivator 7 |
| chr3_+_140981456 | 0.19 |
ENST00000504264.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr4_+_54243917 | 0.19 |
ENST00000507166.1
|
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr12_-_11036844 | 0.18 |
ENST00000428168.2
|
PRH1
|
proline-rich protein HaeIII subfamily 1 |
| chr1_+_40942887 | 0.17 |
ENST00000372706.1
|
ZFP69
|
ZFP69 zinc finger protein |
| chr11_+_63137251 | 0.17 |
ENST00000310969.4
ENST00000279178.3 |
SLC22A9
|
solute carrier family 22 (organic anion transporter), member 9 |
| chrX_-_24690771 | 0.17 |
ENST00000379145.1
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr16_+_20499024 | 0.17 |
ENST00000593357.1
|
AC137056.1
|
Uncharacterized protein; cDNA FLJ34659 fis, clone KIDNE2018863 |
| chr8_+_42195972 | 0.16 |
ENST00000532157.1
ENST00000265421.4 ENST00000520008.1 |
POLB
|
polymerase (DNA directed), beta |
| chr7_-_122840015 | 0.16 |
ENST00000194130.2
|
SLC13A1
|
solute carrier family 13 (sodium/sulfate symporter), member 1 |
| chr12_-_10251539 | 0.16 |
ENST00000420265.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr8_+_24151620 | 0.16 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr2_-_166060571 | 0.15 |
ENST00000360093.3
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr7_-_82792215 | 0.15 |
ENST00000333891.9
ENST00000423517.2 |
PCLO
|
piccolo presynaptic cytomatrix protein |
| chr4_-_69111401 | 0.15 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr12_+_50898881 | 0.15 |
ENST00000301180.5
|
DIP2B
|
DIP2 disco-interacting protein 2 homolog B (Drosophila) |
| chr10_+_35416090 | 0.15 |
ENST00000354759.3
|
CREM
|
cAMP responsive element modulator |
| chr1_-_85930246 | 0.15 |
ENST00000426972.3
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr12_-_10251603 | 0.14 |
ENST00000457018.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr1_+_74701062 | 0.14 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr12_+_95611516 | 0.13 |
ENST00000436874.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr11_+_128563652 | 0.13 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr12_-_10251576 | 0.13 |
ENST00000315330.4
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr1_-_156828810 | 0.13 |
ENST00000368195.3
|
INSRR
|
insulin receptor-related receptor |
| chr1_+_47489240 | 0.13 |
ENST00000371901.3
|
CYP4X1
|
cytochrome P450, family 4, subfamily X, polypeptide 1 |
| chr12_-_10282836 | 0.12 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr21_+_30671690 | 0.12 |
ENST00000399921.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr20_-_45981138 | 0.12 |
ENST00000446994.2
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr12_+_95611569 | 0.12 |
ENST00000261219.6
ENST00000551472.1 ENST00000552821.1 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr19_-_17375541 | 0.12 |
ENST00000252597.3
|
USHBP1
|
Usher syndrome 1C binding protein 1 |
| chr3_-_18466026 | 0.11 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr2_-_162931052 | 0.11 |
ENST00000360534.3
|
DPP4
|
dipeptidyl-peptidase 4 |
| chr1_+_185014496 | 0.11 |
ENST00000367510.3
|
RNF2
|
ring finger protein 2 |
| chr1_-_167883353 | 0.10 |
ENST00000545172.1
|
ADCY10
|
adenylate cyclase 10 (soluble) |
| chr1_-_201476220 | 0.10 |
ENST00000526723.1
ENST00000524951.1 |
CSRP1
|
cysteine and glycine-rich protein 1 |
| chr4_-_40516560 | 0.10 |
ENST00000513473.1
|
RBM47
|
RNA binding motif protein 47 |
| chr1_-_243418650 | 0.09 |
ENST00000522995.1
|
CEP170
|
centrosomal protein 170kDa |
| chr3_-_57583130 | 0.09 |
ENST00000303436.6
|
ARF4
|
ADP-ribosylation factor 4 |
| chr1_-_119532127 | 0.09 |
ENST00000207157.3
|
TBX15
|
T-box 15 |
| chr3_-_19988462 | 0.08 |
ENST00000344838.4
|
EFHB
|
EF-hand domain family, member B |
| chr1_+_185703513 | 0.08 |
ENST00000271588.4
ENST00000367492.2 |
HMCN1
|
hemicentin 1 |
| chr11_+_36317830 | 0.08 |
ENST00000530639.1
|
PRR5L
|
proline rich 5 like |
| chr17_+_67498538 | 0.08 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr11_+_128563948 | 0.08 |
ENST00000534087.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr11_-_18548426 | 0.08 |
ENST00000357193.3
ENST00000536719.1 |
TSG101
|
tumor susceptibility 101 |
| chr2_-_166060382 | 0.07 |
ENST00000409101.3
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr10_-_28270795 | 0.07 |
ENST00000545014.1
|
ARMC4
|
armadillo repeat containing 4 |
| chr9_-_74383302 | 0.07 |
ENST00000377066.5
|
TMEM2
|
transmembrane protein 2 |
| chr1_+_240255166 | 0.07 |
ENST00000319653.9
|
FMN2
|
formin 2 |
| chr5_+_66254698 | 0.07 |
ENST00000405643.1
ENST00000407621.1 ENST00000432426.1 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chrX_+_41192595 | 0.07 |
ENST00000399959.2
|
DDX3X
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr5_+_140235469 | 0.07 |
ENST00000506939.2
ENST00000307360.5 |
PCDHA10
|
protocadherin alpha 10 |
| chr6_-_11779174 | 0.07 |
ENST00000379413.2
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr7_-_107443652 | 0.06 |
ENST00000340010.5
ENST00000422236.2 ENST00000453332.1 |
SLC26A3
|
solute carrier family 26 (anion exchanger), member 3 |
| chr12_-_10282742 | 0.06 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr11_-_85430356 | 0.06 |
ENST00000526999.1
|
SYTL2
|
synaptotagmin-like 2 |
| chr4_+_71457970 | 0.05 |
ENST00000322937.6
|
AMBN
|
ameloblastin (enamel matrix protein) |
| chr2_+_223536428 | 0.05 |
ENST00000446656.3
|
MOGAT1
|
monoacylglycerol O-acyltransferase 1 |
| chr11_-_8964580 | 0.05 |
ENST00000325884.1
|
ASCL3
|
achaete-scute family bHLH transcription factor 3 |
| chr5_+_170288856 | 0.04 |
ENST00000523189.1
|
RANBP17
|
RAN binding protein 17 |
| chr4_+_71458012 | 0.04 |
ENST00000449493.2
|
AMBN
|
ameloblastin (enamel matrix protein) |
| chrY_+_15016013 | 0.04 |
ENST00000360160.4
ENST00000454054.1 |
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr10_+_35415851 | 0.04 |
ENST00000374726.3
|
CREM
|
cAMP responsive element modulator |
| chr6_-_11779014 | 0.03 |
ENST00000229583.5
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr18_+_7946839 | 0.03 |
ENST00000578916.1
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chr2_-_37899323 | 0.03 |
ENST00000295324.3
ENST00000457889.1 |
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr4_-_70725856 | 0.03 |
ENST00000226444.3
|
SULT1E1
|
sulfotransferase family 1E, estrogen-preferring, member 1 |
| chr5_+_38148582 | 0.02 |
ENST00000508853.1
|
CTD-2207A17.1
|
CTD-2207A17.1 |
| chr4_-_87028478 | 0.02 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr4_+_144303093 | 0.02 |
ENST00000505913.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr9_-_85882145 | 0.02 |
ENST00000328788.1
|
FRMD3
|
FERM domain containing 3 |
| chr1_-_243418621 | 0.01 |
ENST00000366544.1
ENST00000366543.1 |
CEP170
|
centrosomal protein 170kDa |
| chr3_+_19988566 | 0.01 |
ENST00000273047.4
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr14_+_52456327 | 0.01 |
ENST00000556760.1
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr19_-_17375527 | 0.01 |
ENST00000431146.2
ENST00000594190.1 |
USHBP1
|
Usher syndrome 1C binding protein 1 |
| chr1_+_228337553 | 0.00 |
ENST00000366714.2
|
GJC2
|
gap junction protein, gamma 2, 47kDa |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX30
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.1 | 1.4 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.5 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.1 | 0.4 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.1 | 0.3 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.2 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.1 | 0.2 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 0.8 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 0.5 | GO:0030421 | defecation(GO:0030421) |
| 0.1 | 1.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.2 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 0.5 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.1 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.3 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.6 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.3 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.1 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.3 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 0.3 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 0.2 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 1.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.3 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.4 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 1.4 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 1.4 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.4 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 0.2 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.1 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.3 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.8 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.3 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.7 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.1 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.5 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.2 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.2 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 1.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.8 | PID ATR PATHWAY | ATR signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.5 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |